Quantitative high-throughput analysis of synthetic genetic interactions in Caenorhabditis elegans by RNA interference
- PMID: 19059334
- PMCID: PMC4443778
- DOI: 10.1016/j.ygeno.2008.11.006
Quantitative high-throughput analysis of synthetic genetic interactions in Caenorhabditis elegans by RNA interference
Abstract
Biological processes are highly dynamic but the current representation of molecular networks is static and largely qualitative. To investigate the dynamic property of genetic networks, a novel quantitative high-throughput method based on RNA interference and capable of calculating the relevance of each interaction, was developed. With this approach, it will be possible to identify not only the components of a network, but also to investigate quantitatively how network and biological processes react to perturbations. As a first application of this method, the genetic interactions of a weak loss-of-function mutation in the gene efl-1/E2F with all the genes of chromosome III were investigated during embryonic development of Caenorhabditis elegans. Fifteen synthetic genetic interactions of efl-1/E2F with the genes of chromosome III were detected, measured and ranked by statistical relevance.
Figures
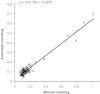

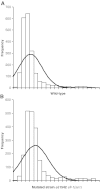
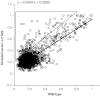
References
-
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. - PubMed
-
- NBrowse. http://www.gnetbrowse.org accessed Dicember 2007
-
- Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38:896–903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

