A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data
- PMID: 19059344
- PMCID: PMC2651152
- DOI: 10.1016/j.neuroimage.2008.10.057
A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data
Abstract
Independent component analysis (ICA) has become an increasingly utilized approach for analyzing brain imaging data. In contrast to the widely used general linear model (GLM) that requires the user to parameterize the data (e.g. the brain's response to stimuli), ICA, by relying upon a general assumption of independence, allows the user to be agnostic regarding the exact form of the response. In addition, ICA is intrinsically a multivariate approach, and hence each component provides a grouping of brain activity into regions that share the same response pattern thus providing a natural measure of functional connectivity. There are a wide variety of ICA approaches that have been proposed, in this paper we focus upon two distinct methods. The first part of this paper reviews the use of ICA for making group inferences from fMRI data. We provide an overview of current approaches for utilizing ICA to make group inferences with a focus upon the group ICA approach implemented in the GIFT software. In the next part of this paper, we provide an overview of the use of ICA to combine or fuse multimodal data. ICA has proven particularly useful for data fusion of multiple tasks or data modalities such as single nucleotide polymorphism (SNP) data or event-related potentials. As demonstrated by a number of examples in this paper, ICA is a powerful and versatile data-driven approach for studying the brain.
Figures





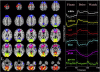
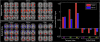


References
-
- Adali T, Novey M, Cardoso JF, Adali T, Novey M, Cardoso JF. Complex ICA using nonlinear functions. IEEE Trans. Signal Process. 2008;59:4356–4544.
-
- Ardnt C. 1996. Information gained by data fusion.
-
- Bach F, Jordan M. Kernel independent component analysis. J. Mach. Learn. Res. 2002;3:1–48.
-
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging. 2004;23(2):137–152. - PubMed
-
- Beckmann CF, Smith SM. Tensorial extensions of independent component analysis for multisubject FMRI analysis. NeuroImage. 2005;25(1):294–311. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

