TGF-beta1 modulates focal adhesion kinase expression in rat intestinal epithelial IEC-6 cells via stimulatory and inhibitory Smad binding elements
- PMID: 19059368
- PMCID: PMC2730956
- DOI: 10.1016/j.bbagrm.2008.11.002
TGF-beta1 modulates focal adhesion kinase expression in rat intestinal epithelial IEC-6 cells via stimulatory and inhibitory Smad binding elements
Abstract
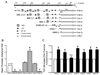
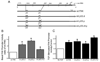
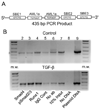
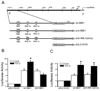
TGF-beta and FAK modulate cell migration, differentiation, proliferation and apoptosis, and TGF-beta promotes FAK transcription in intestinal epithelial cells via Smad-dependent and independent pathways. We utilized a 1320 bp FAK promoter-luciferase construct to characterize basal and TGF-beta-mediated FAK gene transcription in IEC-6 cells. Inhibiting JNK or Akt negated TGF-beta-stimulated promoter activity; ERK inhibition did not block the TGF-beta effect but increased basal activity. Co-transfection with Co-Smad4 enhanced the TGF-beta response while the inhibitory Smad7 abolished it. Serial deletions sequentially removing the four Smad binding elements (SBE) in the 5' untranslated region of the promoter revealed that the two most distal SBE's are positive regulators while SBE3 exerts a negative influence. Mutational deletion of two upstream p53 sites enhanced basal but did not affect TGF-beta-stimulated increases in promoter activity. TGF-beta increased DNA binding of Smad4, phospho-Smad2/3 and Runx1/AML1a to the most distal 435 bp containing 3 SBE and 2 AML1a sites by ChIP assay. However, although point mutation of SBE1 ablated the TGF-beta-mediated rise in SV40-promoter activity, mutation of AML1a sites did not. TGF-beta regulation of FAK transcription reflects a complex interplay between positive and negative non-Smad signals and SBE's, the last independent of p53 or AML1a.
Figures





Similar articles
-
Molecular requirements for induction of CTGF expression by TGF-beta1 in primary osteoblasts.Bone. 2008 May;42(5):871-85. doi: 10.1016/j.bone.2008.01.006. Epub 2008 Jan 26. Bone. 2008. PMID: 18314002 Free PMC article.
-
Activation of extracellular signal-regulated kinase by TGF-beta1 via TbetaRII and Smad7 dependent mechanisms in human bronchial epithelial BEP2D cells.Cell Biol Toxicol. 2007 Mar;23(2):113-28. doi: 10.1007/s10565-006-0097-x. Epub 2006 Nov 9. Cell Biol Toxicol. 2007. PMID: 17096210
-
Smad signaling pathway is a pivotal component of tissue inhibitor of metalloproteinases-3 regulation by transforming growth factor beta in human chondrocytes.Biochim Biophys Acta. 2008 Sep;1783(9):1605-12. doi: 10.1016/j.bbamcr.2008.04.005. Epub 2008 Apr 18. Biochim Biophys Acta. 2008. PMID: 18471442
-
Regulation of elastin promoter by lysyl oxidase and growth factors: cross control of lysyl oxidase on TGF-beta1 effects.Matrix Biol. 2007 Jul;26(6):494-505. doi: 10.1016/j.matbio.2007.02.003. Epub 2007 Feb 27. Matrix Biol. 2007. PMID: 17395448
-
Transforming growth factor-beta stimulates intestinal epithelial focal adhesion kinase synthesis via Smad- and p38-dependent mechanisms.Am J Pathol. 2008 Aug;173(2):385-99. doi: 10.2353/ajpath.2008.070729. Epub 2008 Jun 26. Am J Pathol. 2008. PMID: 18583311 Free PMC article.
Cited by
-
Loss of MLK3 signaling impedes ulcer healing by modulating MAPK signaling in mouse intestinal mucosa.Am J Physiol Gastrointest Liver Physiol. 2012 Oct 15;303(8):G951-60. doi: 10.1152/ajpgi.00158.2012. Epub 2012 Aug 23. Am J Physiol Gastrointest Liver Physiol. 2012. PMID: 22917630 Free PMC article.
-
The role of focal adhesion kinase in the TGF-β-induced myofibroblast transdifferentiation of human Tenon's fibroblasts.Korean J Ophthalmol. 2012 Feb;26(1):45-8. doi: 10.3341/kjo.2012.26.1.45. Epub 2012 Jan 14. Korean J Ophthalmol. 2012. PMID: 22323885 Free PMC article.
-
Role of ERK/mTOR signaling in TGFbeta-modulated focal adhesion kinase mRNA stability and protein synthesis in cultured rat IEC-6 intestinal epithelial cells.Cell Tissue Res. 2009 May;336(2):213-23. doi: 10.1007/s00441-009-0776-z. Epub 2009 Apr 2. Cell Tissue Res. 2009. PMID: 19340459 Free PMC article.
-
Effects and mechanism of irbesartan on tubulointerstitial fibrosis in 5/6 nephrectomized rats.J Huazhong Univ Sci Technolog Med Sci. 2010 Feb;30(1):48-54. doi: 10.1007/s11596-010-0109-1. Epub 2010 Feb 14. J Huazhong Univ Sci Technolog Med Sci. 2010. PMID: 20155455
-
ZINC40099027 Promotes Gastric Mucosal Repair in Ongoing Aspirin-Associated Gastric Injury by Activating Focal Adhesion Kinase.Cells. 2021 Apr 15;10(4):908. doi: 10.3390/cells10040908. Cells. 2021. PMID: 33920786 Free PMC article.
References
-
- Hall PA, Coates PJ, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107(Pt 12):3569–3577. - PubMed
-
- Szabo IL, Pai R, Jones MK, Ehring GR, Kawanaka H, Tarnawski AS. Indomethacin delays gastric restitution: association with the inhibition of focal adhesion kinase and tensin phosphorylation and reduced actin stress fibers. Exp Biol Med (Maywood) 2002;227:412–424. - PubMed
-
- Tarnawski A, Szabo IL, Husain SS, Soreghan B. Regeneration of gastric mucosa during ulcer healing is triggered by growth factors and signal transduction pathways. J Physiol Paris. 2001;95:337–344. - PubMed
-
- Tarnawski AS. Cellular and molecular mechanisms of gastrointestinal ulcer healing. Dig Dis Sci. 2005;50 Suppl 1:S24–S33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

