Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana
- PMID: 19060111
- PMCID: PMC2630444
- DOI: 10.1105/tpc.108.061010
Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana
Abstract
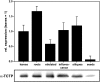
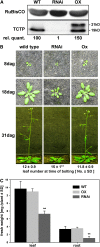
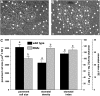
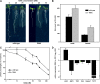
The translationally controlled tumor protein (TCTP) is an important component of the TOR (target of rapamycin) signaling pathway, the major regulator of cell growth in animals and fungi. TCTP acts as the guanine nucleotide exchange factor of the Ras GTPase Rheb that controls TOR activity in Drosophila melanogaster. We therefore examined the role of Arabidopsis thaliana TCTP in planta. Plant TCTPs exhibit distinct sequence differences from nonplant homologs but share the key GTPase binding surface. Green fluorescent protein reporter lines show that Arabidopsis TCTP is expressed throughout plant tissues and developmental stages with increased expression in meristematic and expanding cells. Knockout of TCTP leads to a male gametophytic phenotype with normal pollen formation and germination but impaired pollen tube growth. Silencing of TCTP by RNA interference slows vegetative growth; leaf expansion is reduced because of smaller cell size, lateral root formation is reduced, and root hair development is impaired. Furthermore, these lines show decreased sensitivity to an exogenously applied auxin analog and have elevated levels of endogenous auxin. These results identify TCTP as an important regulator of growth in plants and imply a function of plant TCTP as a mediator of TOR activity similar to that known in nonplant systems.
Figures






Similar articles
-
Identification of Arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes.Plant Cell Physiol. 2013 Oct;54(10):1620-34. doi: 10.1093/pcp/pct106. Epub 2013 Aug 5. Plant Cell Physiol. 2013. PMID: 23921992
-
Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation.Plant Physiol. 2013 Feb;161(2):954-70. doi: 10.1104/pp.112.206029. Epub 2012 Dec 14. Plant Physiol. 2013. PMID: 23370719 Free PMC article.
-
The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis.Plant Cell. 2013 Jun;25(6):2084-101. doi: 10.1105/tpc.113.112359. Epub 2013 Jun 14. Plant Cell. 2013. PMID: 23771893 Free PMC article.
-
Roles of the Translationally Controlled Tumor Protein (TCTP) in Plant Development.Results Probl Cell Differ. 2017;64:149-172. doi: 10.1007/978-3-319-67591-6_7. Results Probl Cell Differ. 2017. PMID: 29149407 Review.
-
Structure-Function Relationship of TCTP.Results Probl Cell Differ. 2017;64:47-68. doi: 10.1007/978-3-319-67591-6_3. Results Probl Cell Differ. 2017. PMID: 29149403 Review.
Cited by
-
Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit.Mol Syst Biol. 2012;8:606. doi: 10.1038/msb.2012.39. Mol Syst Biol. 2012. PMID: 22929616 Free PMC article.
-
Proteomic analysis of tardigrades: towards a better understanding of molecular mechanisms by anhydrobiotic organisms.PLoS One. 2010 Mar 3;5(3):e9502. doi: 10.1371/journal.pone.0009502. PLoS One. 2010. PMID: 20224743 Free PMC article.
-
Monitoring the outside: cell wall-sensing mechanisms.Plant Physiol. 2010 Aug;153(4):1445-52. doi: 10.1104/pp.110.154518. Epub 2010 May 27. Plant Physiol. 2010. PMID: 20508141 Free PMC article. No abstract available.
-
Different effects of night versus day high temperature on rice quality and accumulation profiling of rice grain proteins during grain filling.Plant Cell Rep. 2011 Sep;30(9):1641-59. doi: 10.1007/s00299-011-1074-2. Epub 2011 May 10. Plant Cell Rep. 2011. PMID: 21556707
-
Cyclic AMP deficiency negatively affects cell growth and enhances stress-related responses in tobacco Bright Yellow-2 cells.Plant Mol Biol. 2016 Mar;90(4-5):467-83. doi: 10.1007/s11103-016-0431-5. Epub 2016 Jan 19. Plant Mol Biol. 2016. PMID: 26786166
References
-
- Bai, X., Ma, D., Liu, A., Shen, X., Wang, Q.J., Liu, Y., and Jiang, Y. (2007). Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318 977–980. - PubMed
-
- Baluska, F., Salaj, J., Mathur, J., Braun, M., Jasper, F., Samaj, J., Chua, N.H., Barlow, P.W., and Volkmann, D. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227 618–632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

