Mycobacterium avium genes MAV_5138 and MAV_3679 are transcriptional regulators that play a role in invasion of epithelial cells, in part by their regulation of CipA, a putative surface protein interacting with host cell signaling pathways
- PMID: 19060135
- PMCID: PMC2631991
- DOI: 10.1128/JB.01359-07
Mycobacterium avium genes MAV_5138 and MAV_3679 are transcriptional regulators that play a role in invasion of epithelial cells, in part by their regulation of CipA, a putative surface protein interacting with host cell signaling pathways
Abstract
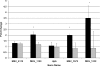
The Mycobacterium avium complex (MAC) is an important group of opportunistic pathogens for birds, cattle, swine, and immunosuppressed humans. Although invasion of epithelial cells lining the intestine is the chief point of entry for these organisms, little is known about the mechanisms by which members of the MAC are taken up by these cells. Studies with M. avium have shown that cytoskeletal rearrangement via activation of the small G-protein Cdc42 is involved and that this activation is regulated in part by the M. avium fadD2 gene. The fadD2 gene indirectly regulates a number of genes upon exposure to HEp-2 cells, including transcriptional regulators, membrane proteins, and secreted proteins. Overexpression of two fadD2-associated regulators (MAV_5138 and MAV_3679) led to increased invasion of HEp-2 cells, as well as altered expression of other genes. The protein product of one of the regulated genes, named CipA, has domains that resemble the PXXP motif of human Piccolo proteins, which bind SH3 domains in proteins involved in the scaffold complex formed during cytoskeletal rearrangement. Although CipA was not detected in the cytoplasm of HEp-2 cells exposed to M. avium, the recombinant protein was shown to be potentially expressed on the surface of Mycobacterium smegmatis incubated with HEp-2 cells and, possibly, to interact with human Cdc42. The interaction was then confirmed by showing that CipA activates Cdc42. These results suggest that members of the M. avium complex have a novel mechanism for activating cytoskeletal rearrangement, prompting uptake by host epithelial cells, and that this mechanism is regulated in part by fadD2, MAV_5138, and MAV_3679.
Figures







References
-
- Alonso-Hearn, M., D. Patel, L. Danelishvili, L. Meunier-Goddik, and L. E. Bermudez. 2008. The Mycobacterium avium subsp. paratuberculosis MAP3464 gene encodes an oxidoreductase involved in invasion of bovine epithelial cells through the activation of host cell Cdc42. Infect. Immun. 76170-178. - PMC - PubMed
-
- Barber, C. E., J. L. Tang, J. X. Feng, M. Q. Pan, T. J. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24555-566. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

