The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex
- PMID: 19060136
- PMCID: PMC2648189
- DOI: 10.1128/JB.01205-08
The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex
Abstract
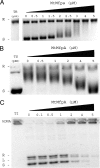
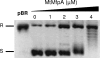
MfpA(Mt) and QnrB4 are two newly characterized pentapeptide repeat proteins (PRPs) that interact with DNA gyrase. The mfpA(Mt) gene is chromosome borne in Mycobacterium tuberculosis, while qnrB4 is plasmid borne in enterobacteria. We expressed and purified the two PRPs and compared their effects on DNA gyrase, taking into account host specificity, i.e., the effect of MfpA(Mt) on M. tuberculosis gyrase and the effect of QnrB4 on Escherichia coli gyrase. Whereas QnrB4 inhibited E. coli gyrase activity only at concentrations higher than 30 microM, MfpA(Mt) inhibited all catalytic reactions of the M. tuberculosis gyrase described for this enzyme (supercoiling, cleavage, relaxation, and decatenation) with a 50% inhibitory concentration of 2 microM. We showed that the D87 residue in GyrA has a major role in the MfpA(Mt)-gyrase interaction, as D87H and D87G substitutions abolished MfpA(Mt) inhibition of M. tuberculosis gyrase catalytic reactions, while A83S modification did not. Since MfpA(Mt) and QnrB4 have been involved in resistance to fluoroquinolones, we measured the inhibition of the quinolone effect in the presence of each PRP. QnrB4 reversed quinolone inhibition of E. coli gyrase at 0.1 microM as described for other Qnr proteins, but MfpA(Mt) did not modify M. tuberculosis gyrase inhibition by fluoroquinolones. Crossover experiments showed that MfpA(Mt) also inhibited E. coli gyrase function, while QnrB4 did not reverse quinolone inhibition of M. tuberculosis gyrase. In conclusion, our in vitro experiments showed that MfpA(Mt) and QnrB4 exhibit opposite effects on DNA gyrase and that these effects are protein and species specific.
Figures






Similar articles
-
Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase.Antimicrob Agents Chemother. 2005 Jan;49(1):118-25. doi: 10.1128/AAC.49.1.118-125.2005. Antimicrob Agents Chemother. 2005. PMID: 15616284 Free PMC article.
-
Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding.J Biol Chem. 2014 May 2;289(18):12300-12. doi: 10.1074/jbc.M113.529164. Epub 2014 Feb 4. J Biol Chem. 2014. PMID: 24497635 Free PMC article.
-
A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA.Science. 2005 Jun 3;308(5727):1480-3. doi: 10.1126/science.1110699. Science. 2005. PMID: 15933203
-
Mechanisms of fluoroquinolone resistance in Mycobacterium tuberculosis.Yi Chuan. 2016 Oct 20;38(10):918-927. doi: 10.16288/j.yczz.16-136. Yi Chuan. 2016. PMID: 27806933 Review.
-
Mycobacterium tuberculosis DNA gyrase as a target for drug discovery.Infect Disord Drug Targets. 2007 Jun;7(2):159-68. doi: 10.2174/187152607781001763. Infect Disord Drug Targets. 2007. PMID: 17970226 Review.
Cited by
-
Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates.J Antimicrob Chemother. 2016 Apr;71(4):882-6. doi: 10.1093/jac/dkv444. Epub 2016 Jan 7. J Antimicrob Chemother. 2016. PMID: 26747096 Free PMC article.
-
Escherichia coli GyrA Tower Domain Interacts with QnrB1 Loop B and Plays an Important Role in QnrB1 Protection from Quinolone Inhibition.Antimicrob Agents Chemother. 2021 Jun 17;65(7):e0040221. doi: 10.1128/AAC.00402-21. Epub 2021 Jun 17. Antimicrob Agents Chemother. 2021. PMID: 33846132 Free PMC article.
-
Interference with KCTD9 inhibits NK cell activation and ameliorates fulminant liver failure in mice.BMC Immunol. 2018 Jun 25;19(1):20. doi: 10.1186/s12865-018-0256-x. BMC Immunol. 2018. PMID: 29940856 Free PMC article.
-
Pentapeptide repeat protein QnrB1 requires ATP hydrolysis to rejuvenate poisoned gyrase complexes.Nucleic Acids Res. 2021 Feb 22;49(3):1581-1596. doi: 10.1093/nar/gkaa1266. Nucleic Acids Res. 2021. PMID: 33434265 Free PMC article.
-
Plasmid-mediated quinolone resistance.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013. Microbiol Spectr. 2014. PMID: 25584197 Free PMC article. Review.
References
-
- Aubry, A., L. M. Fisher, V. Jarlier, and E. Cambau. 2006. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 348158-165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

