Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae
- PMID: 19060153
- PMCID: PMC2632003
- DOI: 10.1128/JB.01465-08
Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae
Abstract
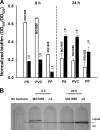
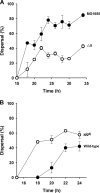
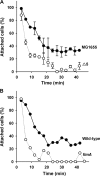
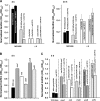
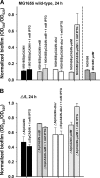
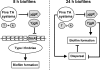
The roles of toxin-antitoxin (TA) systems in bacteria have been debated. Here, the role of five TA systems in regard to biofilm development was investigated (listed as toxin/antitoxin: MazF/MazE, RelE/RelB, ChpB, YoeB/YefM, and YafQ/DinJ). Although these multiple TA systems were reported previously to not impact bacterial fitness, we found that deletion of the five TA systems decreased biofilm formation initially (8 h) on three different surfaces and then increased biofilm formation (24 h) by decreasing biofilm dispersal. Whole-transcriptome profiling revealed that the deletion of the five TA systems induced expression of a single gene, yjgK, which encodes an uncharacterized protein; quantitative real-time PCR (qRT-PCR) confirmed consistent induction of this gene (at 8, 15, and 24 h). Corroborating the complex phenotype seen upon deleting the TA systems, overexpression of YjgK decreased biofilm formation at 8 h and increased biofilm formation at 24 h; deletion of yjgK also affected biofilm formation in the expected manner by increasing biofilm formation after 8 h and decreasing biofilm formation after 24 h. In addition, YjgK significantly reduced biofilm dispersal. Whole-transcriptome profiling revealed YjgK represses fimbria genes at 8 h (corroborated by qRT-PCR and a yeast agglutination assay), which agrees with the decrease in biofilm formation upon deleting the five TA systems at 8 h, as well as that seen upon overexpressing YjgK. Sand column assays confirmed that deleting the five TA systems reduced cell attachment. Furthermore, deletion of each of the five toxins increased biofilm formation at 8 h, and overexpression of the five toxins repressed biofilm formation at 8 h, a result that is opposite that of deleting all five TA systems; this suggests that complex regulation occurs involving the antitoxins. Also, the ability of the global regulator Hha to reduce biofilm formation was dependent on the presence of these TA systems. Hence, we suggest that one role of TA systems is to influence biofilm formation.
Figures


References
-
- Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9773-786. - PubMed
-
- Bayles, K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5721-726. - PubMed
-
- Beloin, C., and J.-M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 1316-19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

