In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70
- PMID: 19060219
- PMCID: PMC2604936
- DOI: 10.1073/pnas.0810851105
In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70
Abstract
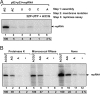
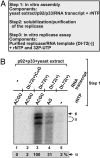
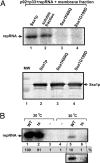
To gain insights into the functions of a viral RNA replicase, we have assembled in vitro and entirely from nonplant sources, a fully functional replicase complex of Tomato bushy stunt virus (TBSV). The formation of the TBSV replicase required two purified recombinant TBSV replication proteins, which were obtained from E. coli, the viral RNA replicon, rATP, rGTP, and a yeast cell-free extract. The in vitro assembly of the replicase took place in the membraneous fraction of the yeast extract, in which the viral replicase-RNA complex became RNase- and proteinase-resistant. The assembly of the replicase complex required the heat shock protein 70 (Hsp70 = yeast Ssa1/2p) present in the soluble fraction of the yeast cell-free extract. The assembled TBSV replicase performed a complete replication cycle, synthesizing RNA complementary to the provided RNA replicon and using the complementary RNA as template to synthesize new TBSV replicon RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Reconstitution of an RNA Virus Replicase in Artificial Giant Unilamellar Vesicles Supports Full Replication and Provides Protection for the Double-Stranded RNA Replication Intermediate.J Virol. 2020 Aug 31;94(18):e00267-20. doi: 10.1128/JVI.00267-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641477 Free PMC article.
-
Activation of Tomato Bushy Stunt Virus RNA-Dependent RNA Polymerase by Cellular Heat Shock Protein 70 Is Enhanced by Phospholipids In Vitro.J Virol. 2015 May;89(10):5714-23. doi: 10.1128/JVI.03711-14. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762742 Free PMC article.
-
A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication.Virology. 2009 Nov 10;394(1):28-38. doi: 10.1016/j.virol.2009.08.003. Epub 2009 Sep 12. Virology. 2009. PMID: 19748649 Free PMC article.
-
Tombusvirus polymerase: Structure and function.Virus Res. 2017 Apr 15;234:74-86. doi: 10.1016/j.virusres.2017.01.012. Epub 2017 Jan 19. Virus Res. 2017. PMID: 28111194 Review.
-
Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus.Adv Virus Res. 2010;76:123-77. doi: 10.1016/S0065-3527(10)76004-8. Epub 2010 Mar 31. Adv Virus Res. 2010. PMID: 20965073 Free PMC article. Review.
Cited by
-
Enrichment of Phosphatidylethanolamine in Viral Replication Compartments via Co-opting the Endosomal Rab5 Small GTPase by a Positive-Strand RNA Virus.PLoS Biol. 2016 Oct 19;14(10):e2000128. doi: 10.1371/journal.pbio.2000128. eCollection 2016 Oct. PLoS Biol. 2016. PMID: 27760128 Free PMC article.
-
The resistance protein Tm-1 inhibits formation of a Tomato mosaic virus replication protein-host membrane protein complex.J Virol. 2013 Jul;87(14):7933-9. doi: 10.1128/JVI.00743-13. Epub 2013 May 8. J Virol. 2013. PMID: 23658455 Free PMC article.
-
A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication.PLoS Pathog. 2011 Dec;7(12):e1002409. doi: 10.1371/journal.ppat.1002409. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174675 Free PMC article.
-
RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes.Proc Natl Acad Sci U S A. 2015 Apr 7;112(14):E1782-91. doi: 10.1073/pnas.1418971112. Epub 2015 Mar 25. Proc Natl Acad Sci U S A. 2015. PMID: 25810252 Free PMC article.
-
Co-opting the fermentation pathway for tombusvirus replication: Compartmentalization of cellular metabolic pathways for rapid ATP generation.PLoS Pathog. 2019 Oct 24;15(10):e1008092. doi: 10.1371/journal.ppat.1008092. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31648290 Free PMC article.
References
-
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338:81–95. - PubMed
-
- Nagy PD, Pogany J. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol Biol. 2008;451:55–68. - PubMed
-
- Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

