The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities
- PMID: 19060898
- PMCID: PMC2615074
- DOI: 10.1038/nsmb.1528
The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities
Abstract
The eukaryotic exosome is a ten-subunit 3' exoribonucleolytic complex responsible for many RNA-processing and RNA-degradation reactions. How the exosome accomplishes this is unknown. Rrp44 (also known as Dis3), a member of the RNase II family of enzymes, is the catalytic subunit of the exosome. We show that the PIN domain of Rrp44 has endoribonucleolytic activity. The PIN domain is preferentially active toward RNA with a 5' phosphate, suggesting coordination of 5' and 3' processing. We also show that the endonuclease activity is important in vivo. Furthermore, the essential exosome subunit Csl4 does not contain any domains that are required for viability, but its zinc-ribbon domain is required for exosome-mediated mRNA decay. These results suggest that specific exosome domains contribute to specific functions, and that different RNAs probably interact with the exosome differently. The combination of an endoRNase and an exoRNase activity seems to be a widespread feature of RNA-degrading machines.
Figures
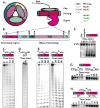

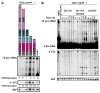

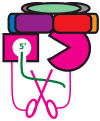
Comment in
-
RNA stability: is it the endo' the world as we know it?Nat Struct Mol Biol. 2009 Jan;16(1):9-10. doi: 10.1038/nsmb0109-9. Nat Struct Mol Biol. 2009. PMID: 19125167 No abstract available.
References
-
- Deutscher MP, Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

