An experimentally derived confidence score for binary protein-protein interactions
- PMID: 19060903
- PMCID: PMC2976677
- DOI: 10.1038/nmeth.1281
An experimentally derived confidence score for binary protein-protein interactions
Abstract
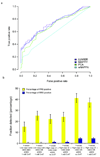
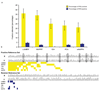
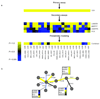
Information on protein-protein interactions is of central importance for many areas of biomedical research. At present no method exists to systematically and experimentally assess the quality of individual interactions reported in interaction mapping experiments. To provide a standardized confidence-scoring method that can be applied to tens of thousands of protein interactions, we have developed an interaction tool kit consisting of four complementary, high-throughput protein interaction assays. We benchmarked these assays against positive and random reference sets consisting of well documented pairs of interacting human proteins and randomly chosen protein pairs, respectively. A logistic regression model was trained using the data from these reference sets to combine the assay outputs and calculate the probability that any newly identified interaction pair is a true biophysical interaction once it has been tested in the tool kit. This general approach will allow a systematic and empirical assignment of confidence scores to all individual protein-protein interactions in interactome networks.
Figures


Comment in
-
Exhaustive benchmarking of the yeast two-hybrid system.Nat Methods. 2010 Sep;7(9):667-8; author reply 668. doi: 10.1038/nmeth0910-667. Nat Methods. 2010. PMID: 20805792 Free PMC article. No abstract available.
References
-
- Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. - PubMed
-
- von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. - PubMed
-
- Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1:349–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015728/ES/NIEHS NIH HHS/United States
- F32 HG 004098/HG/NHGRI NIH HHS/United States
- U01 CA105423/CA/NCI NIH HHS/United States
- R01 ES 015728/ES/NIEHS NIH HHS/United States
- 5U01 CA 105423/CA/NCI NIH HHS/United States
- U54 CA112952/CA/NCI NIH HHS/United States
- F32 HG004098/HG/NHGRI NIH HHS/United States
- P50 HG004233/HG/NHGRI NIH HHS/United States
- 5U54 CA 112952/CA/NCI NIH HHS/United States
- 2R01 HG 001715/HG/NHGRI NIH HHS/United States
- 5P50 HG 004233/HG/NHGRI NIH HHS/United States
- R01 HG003224/HG/NHGRI NIH HHS/United States
- R01 HG 003224/HG/NHGRI NIH HHS/United States
- R01 HG001715/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

