An empirical framework for binary interactome mapping
- PMID: 19060904
- PMCID: PMC2872561
- DOI: 10.1038/nmeth.1280
An empirical framework for binary interactome mapping
Abstract
Several attempts have been made to systematically map protein-protein interaction, or 'interactome', networks. However, it remains difficult to assess the quality and coverage of existing data sets. Here we describe a framework that uses an empirically-based approach to rigorously dissect quality parameters of currently available human interactome maps. Our results indicate that high-throughput yeast two-hybrid (HT-Y2H) interactions for human proteins are more precise than literature-curated interactions supported by a single publication, suggesting that HT-Y2H is suitable to map a significant portion of the human interactome. We estimate that the human interactome contains approximately 130,000 binary interactions, most of which remain to be mapped. Similar to estimates of DNA sequence data quality and genome size early in the Human Genome Project, estimates of protein interaction data quality and interactome size are crucial to establish the magnitude of the task of comprehensive human interactome mapping and to elucidate a path toward this goal.
Figures

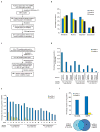

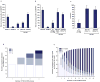
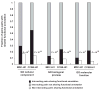
References
-
- Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. - PubMed
-
- Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. - PubMed
-
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA105423/CA/NCI NIH HHS/United States
- 5U01 CA 105423/CA/NCI NIH HHS/United States
- U54 CA112952/CA/NCI NIH HHS/United States
- U01 AI 070499-01/AI/NIAID NIH HHS/United States
- P50 HG004233/HG/NHGRI NIH HHS/United States
- 5U54 CA 112952/CA/NCI NIH HHS/United States
- 2R01 HG 001715/HG/NHGRI NIH HHS/United States
- 5P50 HG 004233/HG/NHGRI NIH HHS/United States
- T32 CA009361/CA/NCI NIH HHS/United States
- T32 CA 09361/CA/NCI NIH HHS/United States
- U56 CA113004/CA/NCI NIH HHS/United States
- U56 CA 113004/CA/NCI NIH HHS/United States
- R01 HG001715/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

