Cancer cells suppress p53 in adjacent fibroblasts
- PMID: 19060923
- PMCID: PMC2727601
- DOI: 10.1038/onc.2008.445
Cancer cells suppress p53 in adjacent fibroblasts
Abstract
The p53 tumor suppressor serves as a crucial barrier against cancer development. In tumor cells and their progenitors, p53 suppresses cancer in a cell-autonomous manner. However, p53 also possesses non-cell-autonomous activities. For example, p53 of stromal fibroblasts can modulate the spectrum of proteins secreted by these cells, rendering their microenvironment less supportive of the survival and spread of adjacent tumor cells. We now report that epithelial tumor cells can suppress p53 induction in neighboring fibroblasts, an effect reproducible by tumor cell-conditioned medium. The ability to suppress fibroblast p53 activation is acquired by epithelial cells in the course of neoplastic transformation. Specifically, stable transduction of immortalized epithelial cells by mutant H-Ras and p53-specific short inhibitory RNA endows them with the ability to quench fibroblast p53 induction. Importantly, human cancer-associated fibroblasts are more susceptible to this suppression than normal fibroblasts. These findings underscore a mechanism whereby epithelial cancer cells may overcome the non-cell-autonomous tumor suppressor function of p53 in stromal fibroblasts.
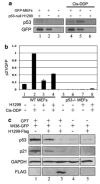
Figures



References
-
- Bar J, Cohen-Noyman E, Geiger B, Oren M. Attenuation of the p53 response to DNA damage by high cell density. Oncogene. 2004;23:2128–2137. - PubMed
-
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, et al. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. - PubMed
-
- Campbell IG, Qiu W, Polyak K, Haviv I. Breast-cancer stromal cells with TP53 mutations. N Engl J Med. 2008;358:1634–1635. author reply 1636. - PubMed
-
- Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264:169–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

