FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease
- PMID: 19061490
- PMCID: PMC2646274
- DOI: 10.1186/gb-2008-9-12-r170
FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease
Abstract
Background: Candidate single nucleotide polymorphisms (SNPs) from genome-wide association studies (GWASs) were often selected for validation based on their functional annotation, which was inadequate and biased. We propose to use the more than 200,000 microarray studies in the Gene Expression Omnibus to systematically prioritize candidate SNPs from GWASs.
Results: We analyzed all human microarray studies from the Gene Expression Omnibus, and calculated the observed frequency of differential expression, which we called differential expression ratio, for every human gene. Analysis conducted in a comprehensive list of curated disease genes revealed a positive association between differential expression ratio values and the likelihood of harboring disease-associated variants. By considering highly differentially expressed genes, we were able to rediscover disease genes with 79% specificity and 37% sensitivity. We successfully distinguished true disease genes from false positives in multiple GWASs for multiple diseases. We then derived a list of functionally interpolating SNPs (fitSNPs) to analyze the top seven loci of Wellcome Trust Case Control Consortium type 1 diabetes mellitus GWASs, rediscovered all type 1 diabetes mellitus genes, and predicted a novel gene (KIAA1109) for an unexplained locus 4q27. We suggest that fitSNPs would work equally well for both Mendelian and complex diseases (being more effective for cancer) and proposed candidate genes to sequence for their association with 597 syndromes with unknown molecular basis.
Conclusions: Our study demonstrates that highly differentially expressed genes are more likely to harbor disease-associated DNA variants. FitSNPs can serve as an effective tool to systematically prioritize candidate SNPs from GWASs.
Figures


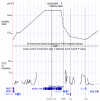
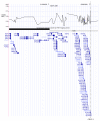
References
-
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. - DOI - PubMed
-
- Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, Steinberg HA, Neto EC, Kleinhanz R, Turner S, Hellerstein MK, Schadt EE, Yandell BS, Kendziorski C, Attie AD. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–716. doi: 10.1101/gr.074914.107. - DOI - PMC - PubMed
-
- Meng H, Vera I, Che N, Wang X, Wang SS, Ingram-Drake L, Schadt EE, Drake TA, Lusis AJ. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc Natl Acad Sci USA. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. - DOI - PMC - PubMed
-
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, Macneil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

