Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues
- PMID: 19061508
- PMCID: PMC2607305
- DOI: 10.1186/1471-2156-9-83
Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues
Abstract
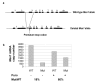
Background: The Nonsense-Mediated mRNA Decay (NMD) pathway detects and degrades mRNAs containing premature termination codons, thereby preventing the accumulation of potentially detrimental truncated proteins. Intertissue variation in the efficiency of this mechanism has been suggested, which could have important implications for the understanding of genotype-phenotype correlations in various genetic disorders. However, compelling evidence in favour of this hypothesis is lacking. Here, we have explored this question by measuring the ratio of mutant versus wild-type Men1 transcripts in thirteen tissues from mice carrying a heterozygous truncating mutation in the ubiquitously expressed Men1 gene.
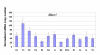
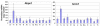
Results: Significant differences were found between two groups of tissues. The first group, which includes testis, ovary, brain and heart, displays a strong decrease of the nonsense transcript (average ratio of 18% of mutant versus wild-type Men1 transcripts, identical to the value measured in murine embryonic fibroblasts). The second group, comprising lung, intestine and thymus, shows much less pronounced NMD (average ratio of 35%). Importantly, the extent of degradation by NMD does not correlate with the expression level of eleven genes encoding proteins involved in NMD or with the expression level of the Men1 gene.
Conclusion: Mouse models are an attractive option to evaluate the efficiency of NMD in multiple mammalian tissues and organs, given that it is much easier to obtain these from a mouse than from a single individual carrying a germline truncating mutation. In this study, we have uncovered in the thirteen different murine tissues that we examined up to a two-fold difference in NMD efficiency.
Figures



References
-
- Tournier I, Raux G, Di Fiore F, Marechal I, Leclerc C, Martin C, Wang Q, Buisine MP, Stoppa-Lyonnet D, Olschwang S, et al. Analysis of the allele-specific expression of the mismatch repair gene MLH1 using a simple DHPLC-Based Method. Hum Mutat. 2004;23:379–384. doi: 10.1002/humu.20008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

