The control of mRNA decapping and P-body formation
- PMID: 19061636
- PMCID: PMC2630519
- DOI: 10.1016/j.molcel.2008.11.001
The control of mRNA decapping and P-body formation
Abstract
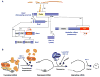
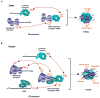
mRNA decapping is a critical step in eukaryotic cytoplasmic mRNA turnover. Cytoplasmic mRNA decapping is catalyzed by Dcp2 in conjunction with its coactivator Dcp1 and is stimulated by decapping enhancer proteins. mRNAs associated with the decapping machinery can assemble into cytoplasmic mRNP granules called processing bodies (PBs). Evidence suggests that PB-associated mRNPs are translationally repressed and can be degraded or stored for subsequent translation. However, whether mRNP assembly into a PB is important for translational repression, decapping, or decay has remained controversial. Here, we discuss the regulation of decapping machinery recruitment to specific mRNPs and how their assembly into PBs is governed by the relative rates of translational repression, mRNP multimerization, and mRNA decay.
Figures

References
-
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

