Morphological appearance manifolds in computational anatomy: groupwise registration and morphological analysis
- PMID: 19061962
- PMCID: PMC2871383
- DOI: 10.1016/j.neuroimage.2008.10.048
Morphological appearance manifolds in computational anatomy: groupwise registration and morphological analysis
Abstract
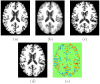
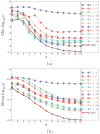
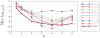
Existing approaches to computational anatomy assume that a perfectly conforming diffeomorphism applied to an anatomy of interest captures its morphological characteristics relative to a template. However, the amount of biological variability in a groupwise analysis renders this task practically impossible, due to the nonexistence of a single template that matches all anatomies in an ensemble, even if such a template is constructed by group averaging procedures. Consequently, anatomical characteristics not captured by the transformation, and which are left out in the residual image, are lost permanently from subsequent analysis, if only properties of the transformation are examined. This paper extends our recent work [Makrogiannis, S., Verma, R., Davatzikos, C., 2007. Anatomical equivalence class: a computational anatomy framework using a lossless shape descriptor. IEEE Trans. Biomed. Imag. 26(4), 619-631] on characterizing subtle morphological variations via a lossless morphological descriptor that takes the residual into account along with the transformation. Since there are infinitely many [transformation, residual] pairs that reconstruct a given anatomy, we treat them as members of an Anatomical Equivalence Class (AEC), thereby forming a manifold embedded in the space spanned by [transformation, residual]. This paper develops a unique and optimal representation of each anatomy that is estimated from the corresponding AECs by solving a global optimization problem. This effectively determines the optimal template and transformation parameters for each individual anatomy, and eliminates respective confounding variation in the data. It, therefore, constitutes the second novelty, in that it represents a group-wise optimal registration strategy that individually adjusts the template and the smoothness of the transformation according to each anatomy. Experimental results support the superiority of our morphological analysis framework over conventional analysis, and demonstrate better diagnostic accuracy.
Figures


















References
-
- Ashburner J, Friston KJ. Voxel-based morphometry – the methods. NeuroImage. 2000;11(6):805–821. - PubMed
-
- Avants BB, Epstein CL, Gee JC. Geodesic image normalization and temporal parameterization in the space of diffeomorphisms. MIAR. 2006:9–16.
-
- Baloch S, Verma R, Davatzikos C. An anatomical equivalence class based joint transformation-residual descriptor for morphological analysis. IPMI. 2007:594–606. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

