Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking
- PMID: 19062174
- PMCID: PMC2909601
- DOI: 10.1002/prot.22309
Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking
Abstract
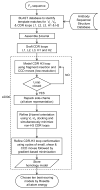
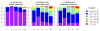
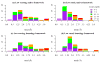
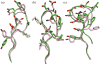
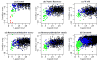
High-resolution homology models are useful in structure-based protein engineering applications, especially when a crystallographic structure is unavailable. Here, we report the development and implementation of RosettaAntibody, a protocol for homology modeling of antibody variable regions. The protocol combines comparative modeling of canonical complementarity determining region (CDR) loop conformations and de novo loop modeling of CDR H3 conformation with simultaneous optimization of V(L)-V(H) rigid-body orientation and CDR backbone and side-chain conformations. The protocol was tested on a benchmark of 54 antibody crystal structures. The median root mean square deviation (rmsd) of the antigen binding pocket comprised of all the CDR residues was 1.5 A with 80% of the targets having an rmsd lower than 2.0 A. The median backbone heavy atom global rmsd of the CDR H3 loop prediction was 1.6, 1.9, 2.4, 3.1, and 6.0 A for very short (4-6 residues), short (7-9), medium (10-11), long (12-14) and very long (17-22) loops, respectively. When the set of ten top-scoring antibody homology models are used in local ensemble docking to antigen, a moderate-to-high accuracy docking prediction was achieved in seven of fifteen targets. This success in computational docking with high-resolution homology models is encouraging, but challenges still remain in modeling antibody structures for sequences with long H3 loops. This first large-scale antibody-antigen docking study using homology models reveals the level of "functional accuracy" of these structural models toward protein engineering applications.
Copyright 2008 Wiley-Liss, Inc.
Figures
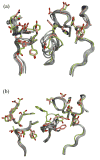
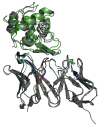
References
-
- Reichert J, Pavlou A. Monoclonal antibodies market. Nature Reviews Drug Discovery. 2004;3(5):383–384. - PubMed
-
- Dufner P, Jermutus L, Minter RR. Harnessing phage and ribosome display for antibody optimisation. Trends in Biotechnology. 2006;24(11):523–529. - PubMed
-
- Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Engineering antibody affinity by yeast surface display. Protein Engineering. 2004;388:348–358. - PubMed
-
- Maynard J, Georgiou G. Antibody engineering. Annu Rev Biomed Eng. 2000;2:339376. - PubMed
-
- Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJM, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, Miller S, Rushe M, Sherman W, Simon K, Van Vlijmen H. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Science. 2006;15(5):949–960. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

