Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control
- PMID: 19062316
- PMCID: PMC2622434
- DOI: 10.1016/j.immuni.2008.10.010
Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control
Abstract
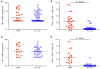
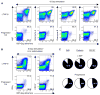
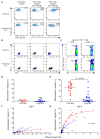
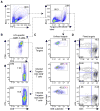
Virus-specific CD8+ T cells probably mediate control over HIV replication in rare individuals, termed long-term nonprogressors (LTNPs) or elite controllers. Despite extensive investigation, the mechanisms responsible for this control remain incompletely understood. We observed that HIV-specific CD8+ T cells of LTNPs persisted at higher frequencies than those of treated progressors with equally low amounts of HIV. Measured on a per-cell basis, HIV-specific CD8+ T cells of LTNPs efficiently eliminated primary autologous HIV-infected CD4+ T cells. This function required lytic granule loading of effectors and delivery of granzyme B to target cells. Defective cytotoxicity of progressor effectors could be restored after treatment with phorbol ester and calcium ionophore. These results establish an effector function and mechanism that clearly segregate with immunologic control of HIV. They also demonstrate that lytic granule contents of memory cells are a critical determinant of cytotoxicity that must be induced for maximal per-cell killing capacity.
Figures


Comment in
-
Elite suppression of HIV-1 replication.Immunity. 2008 Dec 19;29(6):845-7. doi: 10.1016/j.immuni.2008.12.002. Immunity. 2008. PMID: 19100698
References
-
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. - PubMed
-
- Arrode G, Finke JS, Zebroski H, Siegal FP, Steinman RM. CD8+ T cells from most HIV-1-infected patients, even when challenged with mature dendritic cells, lack functional recall memory to HIV gag but not other viruses. Eur J Immunol. 2005;35:159–170. - PubMed
-
- Barber DL, Wherry EJ, Ahmed R. Cutting edge: Rapid in vivo killing by memory CD8 T cells. J Immunol. 2003;171:27–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

