A der(8)t(8;11) chromosome in the Karpas-620 myeloma cell line expresses only cyclin D1: yet both cyclin D1 and MYC are repositioned in close proximity to the 3'IGH enhancer
- PMID: 19064000
- PMCID: PMC2678939
- DOI: 10.1016/j.dnarep.2008.11.010
A der(8)t(8;11) chromosome in the Karpas-620 myeloma cell line expresses only cyclin D1: yet both cyclin D1 and MYC are repositioned in close proximity to the 3'IGH enhancer
Abstract
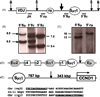
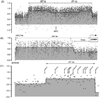
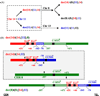
The Karpas-620 human myeloma cell line (HMCL) expresses high levels of Cyclin D1 (CCND1), but has a der(8)t(8;11) and a der(14)t(8;14), and not a conventional t(11;14). Fluorescent in situ hybridization (FISH) and array comparative genomic hybridization (aCGH) studies suggest that der(14)t(11;14) from a primary translocation underwent a secondary translocation with chromosome 8 to generate der(8)t(8;[14];11) and der(14)t(8;[11];14). Both secondary derivatives share extensive identical sequences from chromosomes 8, 11, and 14, including MYC and the 3' IgH enhancers. Der(14), with MYC located approximately 700 kb telomeric to the 3'IGH enhancer, expresses MYC. By contrast, der(8), with both CCND1 and MYC repositioned near a 3'IGH enhancer, expresses CCND1, which is telomeric of the enhancer, but not MYC, which is centromeric to the enhancer. The secondary translocation that dysregulated MYC resulted in extensive regions from both donor chromosomes being transmitted to both derivative chromosomes, suggesting a defect in DNA recombination or repair in the myeloma tumor cell.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

