Whole-genome amplification enables accurate genotyping for microarray-based high-density single nucleotide polymorphism array
- PMID: 19064567
- PMCID: PMC2871542
- DOI: 10.1158/1055-9965.EPI-08-0482
Whole-genome amplification enables accurate genotyping for microarray-based high-density single nucleotide polymorphism array
Abstract
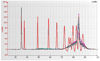
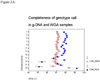
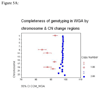
In large-scale genome-wide association studies based on high-density single nucleotide polymorphism (SNP) genotyping array, the quantity and quality of available genomic DNA (gDNA) is a practical problem. We examined the feasibility of using the Multiple Displacement Amplification (MDA) method of whole-genome amplification (WGA) for such a platform. The Affymetrix Early Access Mendel Nsp 250K GeneChip was used for genotyping 224,940 SNPs per sample for 28 DNA samples. We compared the call concordance using 14 gDNA samples and their corresponding 14 WGA samples. The overall mean genotype call rates in gDNA and the corresponding WGA samples were comparable at 97.07% [95% confidence interval (CI), 96.17-97.97] versus 97.77% (95% CI, 97.26-98.28; P = 0.154), respectively. Reproducibility of the platform, calculated as concordance in duplicate samples, was 99.45%. Overall genotypes for 97.74% (95% CI, 97.03-98.44) of SNPs were concordant between gDNA and WGA samples. When the analysis was restricted to well-performing SNPs (successful genotyping in gDNA and WGA in >90% of samples), 99.11% (95% CI, 98.80-99.42) of the SNPs, on average, were concordant, and overall a SNP showed a discordant call in 0.92% (95% CI, 0.90-0.94) of paired samples. In a pair of gDNA and WGA DNA, similar concordance was reproducible on Illumina's Infinium 610 Quad platform as well. Although copy number analysis revealed a total of seven small telomeric regions in six chromosomes with loss of copy number, the estimated genome representation was 99.29%. In conclusion, our study confirms that high-density oligonucleotide array-based genotyping can yield reproducible data and MDA-WGA DNA products can be effectively used for genome-wide SNP genotyping analysis.
Figures








References
-
- Harty LC, Garcia-Closas M, Rothman N, Reid YA, Tucker MA, Hartge P. Collection of buccal cell DNA using treated cards. Cancer Epidemiol Biomarkers Prev. 2000;9:501–506. - PubMed
-
- Packer RJ, Bolton BJ. A Laboratory Handbook: Immortalization of B-Lymphocyte by Epstein-Barr Virus. Academic Press; 1998. Cell Biology.
-
- Little SE, Vuononvirta R, Reis-Filho JS, et al. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics. 2006;87:298–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

