Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement
- PMID: 19066219
- PMCID: PMC2605001
- DOI: 10.1073/pnas.0807826105
Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement
Abstract
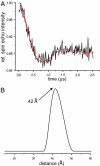
alpha-Synuclein is known to play a causative role in Parkinson disease. Although its physiological functions are not fully understood, alpha-synuclein has been shown to interact with synaptic vesicles and modulate neurotransmitter release. However, the structure of its physiologically relevant membrane-bound state remains unknown. Here we developed a site-directed spin labeling and EPR-based approach for determining the structure of alpha-synuclein bound to a lipid bilayer. Continuous-wave EPR was used to assign local secondary structure and to determine the membrane immersion depth of lipid-exposed residues, whereas pulsed EPR was used to map long-range distances. The structure of alpha-synuclein was built and refined by using simulated annealing molecular dynamics restrained by the immersion depths and distances. We found that alpha-synuclein forms an extended, curved alpha-helical structure that is over 90 aa in length. The monomeric helix has a superhelical twist similar to that of right-handed coiled-coils which, like alpha-synuclein, contain 11-aa repeats, but which are soluble, oligomeric proteins (rmsd = 0.82 A). The alpha-synuclein helix extends parallel to the curved membrane in a manner that allows conserved Lys and Glu residues to interact with the zwitterionic headgroups, while uncharged residues penetrate into the acyl chain region. This structural arrangement is significantly different from that of alpha-synuclein in the presence of the commonly used membrane-mimetic detergent SDS, which induces the formation of two antiparallel helices. Our structural analysis emphasizes the importance of studying membrane protein structure in a bilayer environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



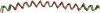
References
-
- Cole NB, et al. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein alpha-synuclein. J Biol Chem. 2002;277:6344–6352. - PubMed
-
- Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–678. - PubMed
-
- Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: Dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. - PubMed
-
- Zhu M, Li J, Fink AL. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J Biol Chem. 2003;278:40186–40197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

