Measurement of imino 1H-1H residual dipolar couplings in RNA
- PMID: 19067179
- PMCID: PMC2846714
- DOI: 10.1007/s10858-008-9293-8
Measurement of imino 1H-1H residual dipolar couplings in RNA
Abstract
Imino (1)H-(15)N residual dipolar couplings (RDCs) provide additional structural information that complements standard (1)H-(1)H NOEs leading to improvements in both the local and global structure of RNAs. Here, we report measurement of imino (1)H-(1)H RDCs for the Iron Responsive Element (IRE) RNA and native E. coli tRNA(Val) using a BEST-Jcomp-HMQC2 experiment. (1)H-(1)H RDCs are observed between the imino protons in G-U wobble base pairs and between imino protons on neighboring base pairs in both RNAs. These imino (1)H-(1)H RDCs complement standard (1)H-(15)N RDCs because the (1)H-(1)H vectors generally point along the helical axis, roughly perpendicular to (1)H-(15)N RDCs. The use of longitudinal relaxation enhancement increased the signal-to-noise of the spectra by ~3.5-fold over the standard experiment. The ability to measure imino (1)H-(1)H RDCs offers a new restraint, which can be used in NMR domain orientation and structural studies of RNAs.
Figures
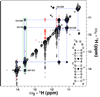


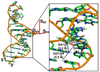

References
-
- Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi A. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997;274:72–83. - PubMed
-
- Al-Hashimi HM, Gosser Y, Gorin A, Hu WD, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. - PubMed
-
- Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. - PubMed
-
- Bondensgaard K, Mollova ET, Pardi A. The global conformation of the hammerhead ribozyme determined using residual dipolar couplings. Biochemistry. 2002;41:11532–11542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

