Cell-to-cell signalling during pathogenesis
- PMID: 19068097
- PMCID: PMC2786497
- DOI: 10.1111/j.1462-5822.2008.01272.x
Cell-to-cell signalling during pathogenesis
Abstract
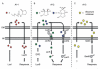
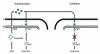
Pathogenic bacteria produce virulence factors only when they sense they are in a location in which the energy required for pathogenesis is warranted. One environmental factor monitored by pathogens is population density, either of its own population or of the population of a host's endogenous flora. To date, four systems have been described which allow pathogens to regulate virulence genes in a population-dependent manner. These systems are found in both Gram-positive and Gram-negative cells and utilize various mechanisms to control gene regulation at a transcriptional level, but they all have one feature in common: they detect autoinducers released by cells belonging to either the pathogen's population or that of the host's flora. This article explores the role of these four signalling systems in bacterial communities and how pathogens use these systems to control genes required during host invasion and infection.
Figures
References
-
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. - PubMed
-
- Coulthurst SJ, Williamson NR, Harris AK, Spring DR, Salmond GP. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology. 2006;152:1899–1911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

