Epigenomic consequences of immortalized plant cell suspension culture
- PMID: 19071958
- PMCID: PMC2596858
- DOI: 10.1371/journal.pbio.0060302
Epigenomic consequences of immortalized plant cell suspension culture
Abstract
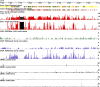
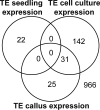
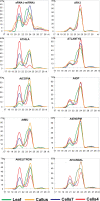
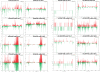
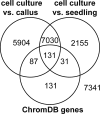
Plant cells grown in culture exhibit genetic and epigenetic instability. Using a combination of chromatin immunoprecipitation and DNA methylation profiling on tiling microarrays, we have mapped the location and abundance of histone and DNA modifications in a continuously proliferating, dedifferentiated cell suspension culture of Arabidopsis. We have found that euchromatin becomes hypermethylated in culture and that a small percentage of the hypermethylated genes become associated with heterochromatic marks. In contrast, the heterochromatin undergoes dramatic and very precise DNA hypomethylation with transcriptional activation of specific transposable elements (TEs) in culture. High throughput sequencing of small interfering RNA (siRNA) revealed that TEs activated in culture have increased levels of 21-nucleotide (nt) siRNA, sometimes at the expense of the 24-nt siRNA class. In contrast, TEs that remain silent, which match the predominant 24-nt siRNA class, do not change significantly in their siRNA profiles. These results implicate RNA interference and chromatin modification in epigenetic restructuring of the genome following the activation of TEs in immortalized cell culture.
Conflict of interest statement
Figures

References
-
- Lee M, Phillips RL. The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:413–437.
-
- Osborne DJ, Mcmanus MT. Flexibility and commitment in plant cells during development. Curr Top Dev Biol. 1986;20:383–396. - PubMed
-
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;54:118–130. - PubMed
-
- Meins F. Habituation: heritable variation in the requirement of cultured plant-cells for hormones. Annu Rev Genet. 1989;23:395–408. - PubMed
-
- Meins F, Foster R. Reversible, cell-heritable changes during the development of tobacco pith tissues. Dev Biol. 1985;108:1–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

