Analytical performance of immobilized pronase for glycopeptide footprinting and implications for surpassing reductionist glycoproteomics
- PMID: 19072223
- PMCID: PMC2637306
- DOI: 10.1021/pr800708h
Analytical performance of immobilized pronase for glycopeptide footprinting and implications for surpassing reductionist glycoproteomics
Abstract
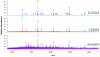
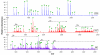
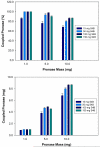
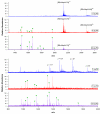
A fully developed understanding of protein glycosylation requires characterization of the modifying oligosaccharides, elucidation of their covalent attachment sites, and determination of the glycan heterogeneity at specific sites. Considering the complexity inherent to protein glycosylation, establishing these features for even a single protein can present an imposing challenge. To meet the demands of glycoproteomics, the capability to screen far more complex systems of glycosylated proteins must be developed. Although the proteome wide examination of carbohydrate modification has become an area of keen interest, the intricacy of protein glycosylation has frustrated the progress of large-scale, systems oriented research on site-specific protein-glycan relationships. Indeed, the analytical obstacles in this area have been more instrumental in shaping the current glycoproteomic paradigm than have the diverse functional roles and ubiquitous nature of glycans. This report describes the ongoing development and analytically salient features of bead immobilized pronase for glycosylation site footprinting. The present work bears on the ultimate goal of providing analytical tools capable of addressing the diversity of protein glycosylation in a more comprehensive and efficient manner. In particular, this approach has been assessed with respect to reproducibility, sensitivity, and tolerance to sample complexity. The efficiency of pronase immobilization, attainable pronase loading density, and the corresponding effects on glycoprotein digestion rate were also evaluated. In addition to being highly reproducible, the immobilized enzymes retained a high degree of proteolytic activity after repeat usage for up to 6 weeks. This method also afforded a low level of chemical background and provided favorable levels of sensitivity with respect to traditional glycoproteomic strategies. Thus, the application of immobilized pronase shows potential to contribute to the advancement of more comprehensive glycoproteomic research methods that are capable of providing site-specific glycosylation and microheterogeneity information across many proteins.
Figures

References
-
- Dwek RA. Glycobiology: toward understanding the function of sugars. Chemical Reviews. 1996;96:683–720. - PubMed
-
- Rudd PM, Dwek RA. Glycosylation: heterogeneity and the 3D structure of proteins. Critical Reviews in Biochemistry and Molecular Biology. 1997;32:1–100. - PubMed
-
- Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43r–56r. - PubMed
-
- Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography-mass spectrometry. Journal of Chromatography B. 2005;825:124–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

