Characterization of a protective Escherichia coli-expressed Plasmodium falciparum merozoite surface protein 3 indicates a non-linear, multi-domain structure
- PMID: 19073223
- PMCID: PMC3633458
- DOI: 10.1016/j.molbiopara.2008.11.006
Characterization of a protective Escherichia coli-expressed Plasmodium falciparum merozoite surface protein 3 indicates a non-linear, multi-domain structure
Abstract
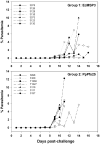
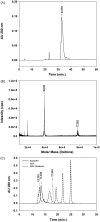
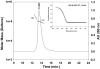
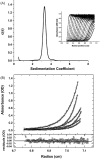
Immunization with a recombinant yeast-expressed Plasmodium falciparum merozoite surface protein 3 (MSP3) protected Aotus nancymai monkeys against a virulent challenge infection. Unfortunately, the production process for this yeast-expressed material was not optimal for human trials. In an effort to produce a recombinant MSP3 protein in a scaleable manner, we expressed and purified near-full-length MSP3 in Escherichia coli (EcMSP3). Purified EcMSP3 formed non-globular dimers as determined by analytical size-exclusion HPLC with in-line multi-angle light scatter and quasi-elastic light scatter detection and velocity sedimentation (R(h) 7.6+/-0.2nm and 6.9nm, respectively). Evaluation by high-resolution atomic force microscopy revealed non-linear asymmetric structures, with beaded domains and flexible loops that were recognized predominantly as dimers, although monomers and larger multimers were observed. The beaded substructure corresponds to predicted structural domains, which explains the velocity sedimentation results and improves the conceptual model of the protein. Vaccination with EcMSP3 in Freund's adjuvant-induced antibodies that recognized native MSP3 in parasitized erythrocytes by an immunofluorescence assay and gave delayed time to treatment in a group of Aotus monkeys in a virulent challenge infection with the FVO strain of P. falciparum. Three of the seven monkeys vaccinated with EcMSP3 had low peak parasitemias. EcMSP3, which likely mimics the native MSP3 structure located on the merozoite surface, is a viable candidate for inclusion in a multi-component malaria vaccine.
Figures



References
-
- McColl DJ, Silva A, Foley M, et al. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994;68(1):53–67. - PubMed
-
- Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, et al. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood. 1994;84(5):1594–602. - PubMed
-
- Singh S, Soe S, Mejia JP, et al. Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J Infect Dis. 2004;190(5):1010–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

