Interactions of HIV-1 inhibitory peptide T20 with the gp41 N-HR coiled coil
- PMID: 19073602
- PMCID: PMC2635040
- DOI: 10.1074/jbc.M809269200
Interactions of HIV-1 inhibitory peptide T20 with the gp41 N-HR coiled coil
Abstract
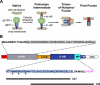
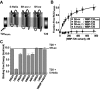
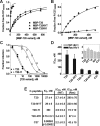
Cellular entry of human immunodeficiency virus type 1 (HIV-1) involves fusion of viral and cellular membranes and is mediated by structural transitions in viral glycoprotein gp41. The antiviral C-peptide T20 targets the gp41 N-terminal heptad repeat region (N-HR), blocking gp41 conformational changes essential for the entry process. To probe the T20 structure-activity relationship, we engineered a molecular mimic of the entire gp41 N-HR coiled coil using the 5-Helix design strategy. T20 bound this artificial protein (denoted 5H-ex) with nanomolar affinity (K(D) = 30 nm), close to its IC50 concentration (approximately 3 nm) but much weaker than the affinity of a related inhibitory C-peptide C37 (K(D) = 0.0007 nm). T20/C37 competitive binding assays confirmed that T20 interacts with the hydrophobic groove on the surface of the N-HR coiled coil outside of a deep pocket region crucial for C37 binding. We used 5H-ex to investigate how the T20 N and C termini contributed to the inhibitor binding activity. Mutating three aromatic residues at the T20 C terminus (WNWF --> ANAA) had no effect on affinity, suggesting that these amino acids do not participate in T20 binding to the gp41 N-HR. The results support recent evidence pointing to a different role for these residues in T20 inhibition (Peisajovich, S. G., Gallo, S. A., Blumenthal, R., and Shai, Y. (2003) J. Biol. Chem. 278, 21012-21017; Liu, S., Jing, W., Cheung, B., Lu, H., Sun, J., Yan, X., Niu, J., Farmar, J., Wu, S., and Jiang, S. (2007) J. Biol. Chem. 282, 9612-9620). By contrast, mutations near the T20 N terminus substantially influenced inhibitor binding strength. When Ile was substituted for Thr in the second T20 position, a 40-fold increase in binding affinity was measured (K(D) = 0.75 nm). The effect of this affinity enhancement on T20 inhibitory potency varied among different viral strains. The original T20 and the higher affinity T20 variant had similar potency against wild type HIV-1. However, the higher affinity T20 variant was significantly more potent against T20-resistant virus. The findings suggest that other factors in addition to binding affinity play a role in limiting T20 potency. As a mimetic of the complete gp41 N-HR coiled coil region, 5H-ex will be a useful tool to further elucidate mechanistic profiles of C-peptide inhibitors.
Figures



References
-
- Peisajovich, S. G., Gallo, S. A., Blumenthal, R., and Shai, Y. (2003) J. Biol. Chem. 278 21012-21017 - PubMed
-
- Liu, S., Jing, W., Cheung, B., Lu, H., Sun, J., Yan, X., Niu, J., Farmar, J., Wu, S., and Jiang, S. (2007) J. Biol. Chem. 282 9612-9620 - PubMed
-
- Fenouillet, E., and Jones, I. M. (1995) J. Gen. Virol. 76 1509-1514 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

