Origin and biology of simian immunodeficiency virus in wild-living western gorillas
- PMID: 19073717
- PMCID: PMC2643789
- DOI: 10.1128/JVI.02311-08
Origin and biology of simian immunodeficiency virus in wild-living western gorillas
Abstract
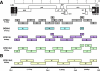
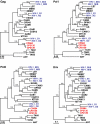
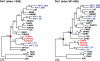
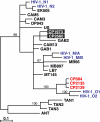
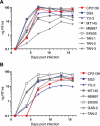
Western lowland gorillas (Gorilla gorilla gorilla) are infected with a simian immunodeficiency virus (SIVgor) that is closely related to chimpanzee and human immunodeficiency viruses (SIVcpz and HIV-1, respectively) in west central Africa. Although existing data suggest a chimpanzee origin for SIVgor, a paucity of available sequences has precluded definitive conclusions. Here, we report the molecular characterization of one partial (BQ664) and three full-length (CP684, CP2135, and CP2139) SIVgor genomes amplified from fecal RNAs of wild-living gorillas at two field sites in Cameroon. Phylogenetic analyses showed that all SIVgor strains clustered together, forming a monophyletic lineage throughout their genomes. Interestingly, the closest relatives of SIVgor were not SIVcpzPtt strains from west central African chimpanzees (Pan troglodytes troglodytes) but human viruses belonging to HIV-1 group O. In trees derived from most genomic regions, SIVgor and HIV-1 group O formed a sister clade to the SIVcpzPtt lineage. However, in a tree derived from 5' pol sequences ( approximately 900 bp), SIVgor and HIV-1 group O fell within the SIVcpzPtt radiation. The latter was due to two SIVcpzPtt strains that contained mosaic pol sequences, pointing to the existence of a divergent SIVcpzPtt lineage that gave rise to SIVgor and HIV-1 group O. Gorillas appear to have acquired this lineage at least 100 to 200 years ago. To examine the biological properties of SIVgor, we synthesized a full-length provirus from fecal consensus sequences. Transfection of the resulting clone (CP2139.287) into 293T cells yielded infectious virus that replicated efficiently in both human and chimpanzee CD4(+) T cells and used CCR5 as the coreceptor for viral entry. Together, these results provide strong evidence that P. t. troglodytes apes were the source of SIVgor. These same apes may also have spawned the group O epidemic; however, the possibility that gorillas served as an intermediary host cannot be excluded.
Figures




References
-
- Akaike, H. 1974. A new look at statistical model identification. IEEE Trans. Automat. Control 19716-723.
-
- Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 965698-5703. - PMC - PubMed
-
- Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Boston, MA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

