Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen
- PMID: 19073746
- PMCID: PMC2643754
- DOI: 10.1128/JVI.02011-08
Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen
Abstract
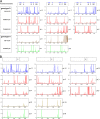
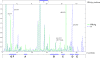
Direct population sequencing and reverse hybridization (line probe assay [LiPA])-based methods are the most common methods for detecting hepatitis B virus (HBV) drug resistance mutations, although only mutations present in viral quasispecies with a prevalence of > or =20% can be detected by sequencing, and only known mutations are detected by LiPA. Massively parallel ultradeep pyrosequencing (UDPS; GS FLX platform) was used to analyze HBV quasispecies in reverse transcriptase (RT) and hepatitis B S antigen (HBsAg) from five drug-naive patients and eight drug-resistant patients. Eight primer pairs were used to obtain partially overlapping amplicons, covering the RT gene from codons 1 to 288 and the complete overlapping HBsAg sequence. A 1% mutation frequency was selected as the cutoff based on an error rate estimated on plasmid DNA. This technology enabled simultaneous analysis of between 2,852 and 18,016 clonally amplified fragments from each patient. The results indicate that UDPS has a relative sensitivity much higher than both direct sequencing and LiPA. In addition, the UDPS results are quantitative, allowing establishment of the relative frequency of both known mutations and novel substitutions. Some of the detected RT substitutions led to changes also in HBsAg. On the whole, genotype D presented a higher heterogeneity than genotype A. Considering the high quantity of information that can be provided by a single test from one patient, the short turnaround time, the information on substitution frequency, and the detection of rare variants, there are strong advantages conferred by UDPS, and the new method could play a relevant role in the clinical management of HBV infection and therapy.
Figures
References
-
- Agresti, A., and B. A. Coull. 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52119-126.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Colonno, R. J., R. Rose, C. J. Baldick, S. Levine, K. Pokornowski, C. F. Yu, A. Walsh, J. Fang, M. Hsu, C. Mazzucco, B. Eggers, S. Zhang, M. Plym, K. Klesczewski, and D. J. Tenney. 2006. Entecavir resistance is rare in nucleoside naïve patients with hepatitis B. Hepatology 441656-1665. - PubMed
-
- De Maddalena, C., C. Giambelli, E. Tanzi, D. Colzani, M. Schiavini, L. Milazzo, F. Bernini, E. Ebranati, A. Cargnel, R. Bruno, M. Galli, and G. Zehender. 2007. High level of genetic heterogeneity in S and P genes of genotype D hepatitis B virus. Virology 365113-124. - PubMed
-
- Dienstag, J. L., R. D. Goldin, E. J. Heathcote, H. W. Hann, M. Woessner, S. L. Stephenson, S. Gardner, D. F. Gray, and E. R. Schiff. 2003. Histological outcome during long-term lamivudine therapy. Gastroenterology 124105-117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

