Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins
- PMID: 19073890
- PMCID: PMC2633399
- DOI: 10.1091/mbc.e08-02-0140
Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins
Abstract
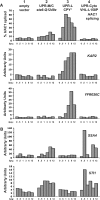
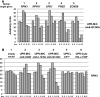
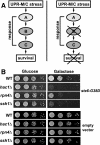
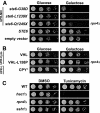
ER quality control (ERQC) prevents the exit of misfolded secretory and membrane proteins from the ER. A critical aspect of ERQC is a transcriptional response called the unfolded protein response (UPR), which up-regulates genes that enable cells to cope with misfolded, ER-retained proteins. In this study, we compare the transcriptional responses in yeast resulting from the acute expression of misfolded proteins residing in three different cellular compartments (the ER lumen, membrane, and cytosol), and find that each elicits a distinct transcriptional response. The classical UPR response, here-designated UPR-L, is induced by the ER lumenal misfolded protein, CPY*. The UPR-Cyto response is induced by the cytosolic protein, VHL-L158P, and is characterized by a rapid, transient induction of cytosolic chaperones similar to the heat-shock response. In contrast, the misfolded membrane protein with a cystolic lesion, Ste6p*, elicits a unique response designated UPR-M/C, characterized by the modest induction of >20 genes regulated by Rpn4p, an activator of proteasomal genes. Independently, we identified several genes required for yeast viability during UPR-M/C stress, but not UPR-L or UPR-Cyto stress. Among these is RPN4, highlighting the importance of the Rpn4p-dependent response in tolerating UPR-M/C stress. Further analysis suggests the requirement for Rpn4p reflects severe impairment of the proteasome by UPR-M/C stress.
Figures


References
-
- Apodaca J., Kim I., Rao H. Cellular tolerance of prion protein PrP in yeast involves proteolysis and the unfolded protein response. Biochem. Biophys. Res. Commun. 2006;347:319–326. - PubMed
-
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
-
- Baker B. M., Tortorella D. Dislocation of an endoplasmic reticulum membrane glycoprotein involves the formation of partially dislocated ubiquitinated polypeptides. J. Biol. Chem. 2007;282:26845–26856. - PubMed
-
- Bence N. F., Sampat R. M., Kopito R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

