One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome
- PMID: 19073939
- PMCID: PMC2600582
- DOI: 10.1073/pnas.0811011106
One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome
Abstract
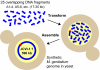
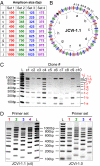
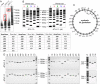
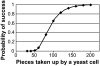
We previously reported assembly and cloning of the synthetic Mycoplasma genitalium JCVI-1.0 genome in the yeast Saccharomyces cerevisiae by recombination of six overlapping DNA fragments to produce a 592-kb circle. Here we extend this approach by demonstrating assembly of the synthetic genome from 25 overlapping fragments in a single step. The use of yeast recombination greatly simplifies the assembly of large DNA molecules from both synthetic and natural fragments.
Conflict of interest statement
Conflict of interest statement: C.A.H., H.O.S., and J.C.V. have a financial interest in Synthetic Genomics, Inc (SGI). The research reported in this paper is in the field in which SGI is developing products. SGI has funded the research reported in the paper.
Figures

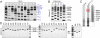
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

