RAD54 controls access to the invading 3'-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae
- PMID: 19074197
- PMCID: PMC2632917
- DOI: 10.1093/nar/gkn980
RAD54 controls access to the invading 3'-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae
Abstract
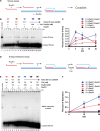
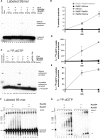
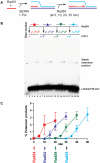
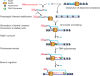
Rad51 is a key protein in homologous recombination performing homology search and DNA strand invasion. After DNA strand exchange Rad51 protein is stuck on the double-stranded heteroduplex DNA product of DNA strand invasion. This is a problem, because DNA polymerase requires access to the invading 3'-OH end to initiate DNA synthesis. Here we show that, the Saccharomyces cerevisiae dsDNA motor protein Rad54 solves this problem by dissociating yeast Rad51 protein bound to the heteroduplex DNA after DNA strand invasion. The reaction required species-specific interaction between both proteins and the ATPase activity of Rad54 protein. This mechanism rationalizes the in vivo requirement of Rad54 protein for the turnover of Rad51 foci and explains the observed dependence of the transition from homologous pairing to DNA synthesis on Rad54 protein in vegetative and meiotic yeast cells.
Figures


References
-
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. - PubMed
-
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. - PubMed
-
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. - PubMed
-
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:570–603. - PubMed
-
- Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

