Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome
- PMID: 19074258
- PMCID: PMC2634942
- DOI: 10.1073/pnas.0805830106
Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome
Abstract
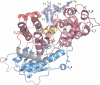
DNA photolyases and cryptochromes (cry) form a family of flavoproteins that use light energy in the blue/UV-A region for the repair of UV-induced DNA lesions or for signaling, respectively. Very recently, it was shown that members of the DASH cryptochrome subclade repair specifically cyclobutane pyrimidine dimers (CPDs) in UV-damaged single-stranded DNA. Here, we report the crystal structure of Arabidopsis cryptochrome 3 with an in-situ-repaired CPD substrate in single-stranded DNA. The structure shows a binding mode similar to that of conventional DNA photolyases. Furthermore, CPD lesions in double-stranded DNA are bound and repaired with similar efficiency as in single-stranded DNA if the CPD lesion is present in a loop structure. Together, these data reveal that DASH cryptochromes catalyze light-driven DNA repair like conventional photolyases but lack an efficient flipping mechanism for interaction with CPD lesions within duplex DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

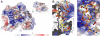

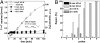
References
-
- Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. - PubMed
-
- Essen LO. Photolyases and cryptochromes: Common mechanisms of DNA repair and light-driven signaling? Curr Opin Struct Biol. 2006;16:51–59. - PubMed
-
- Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. - PubMed
-
- Tamada T, et al. Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol. 1997;4:887–891. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

