Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites
- PMID: 19074313
- PMCID: PMC2757552
- DOI: 10.1126/science.1163183
Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites
Abstract
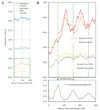
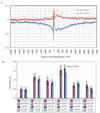
Might DNA sequence variation reflect germline genetic activity and underlying chromatin structure? We investigated this question using medaka (Japanese killifish, Oryzias latipes), by comparing the genomic sequences of two strains (Hd-rR and HNI) and by mapping approximately 37.3 million nucleosome cores from Hd-rR blastulae and 11,654 representative transcription start sites from six embryonic stages. We observed a distinctive approximately 200-base pair (bp) periodic pattern of genetic variation downstream of transcription start sites; the rate of insertions and deletions longer than 1 bp peaked at positions of approximately +200, +400, and +600 bp, whereas the point mutation rate showed corresponding valleys. This approximately 200-bp periodicity was correlated with the chromatin structure, with nucleosome occupancy minimized at positions 0, +200, +400, and +600 bp. These data exemplify the potential for genetic activity (transcription) and chromatin structure to contribute to molding the DNA sequence on an evolutionary time scale.
Figures

Comment in
-
Molecular biology. The structure of change.Science. 2009 Jan 16;323(5912):347-8. doi: 10.1126/science.1169408. Science. 2009. PMID: 19150834 No abstract available.
References
-
- Svejstrup J. Nat. Rev. Mol. Cell Biol. 2002;3:21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

