Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri
- PMID: 19074387
- PMCID: PMC2655524
- DOI: 10.1128/JB.01547-08
Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri
Abstract
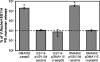
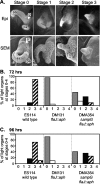
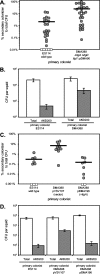
The light-organ symbiont Vibrio fischeri releases N-acetylglucosaminyl-1,6-anhydro-N-acetylmuramylalanyl-gamma-glutamyldiaminopimelylalanine, a disaccharide-tetrapeptide component of peptidoglycan that is referred to here as "PG monomer." In contrast, most gram-negative bacteria recycle PG monomer efficiently, and it does not accumulate extracellularly. PG monomer can stimulate normal light-organ morphogenesis in the host squid Euprymna scolopes, resulting in regression of ciliated appendages similar to that triggered by infection with V. fischeri. We examined whether the net release of PG monomers by V. fischeri resulted from lytic transglycosylase activity or from defects in AmpG, the permease through which PG monomers enter the cytoplasm for recycling. An ampG mutant displayed a 100-fold increase in net PG monomer release, indicating that AmpG is functional. The ampG mutation also conferred the uncharacteristic ability to induce light-organ morphogenesis even when placed in a nonmotile flaJ mutant that cannot infect the light-organ crypts. We targeted five potential lytic transglycosylase genes singly and in specific combinations to assess their role in PG monomer release. Combinations of mutations in ltgA, ltgD, and ltgY decreased net PG monomer release, and a triple mutant lacking all three of these genes had little to no accumulation of PG monomers in culture supernatants. This mutant colonized the host as well as the wild type did; however, the mutant-infected squid were more prone to later superinfection by a second V. fischeri strain. We propose that the lack of PG monomer release by this mutant results in less regression of the infection-promoting ciliated appendages, leading to this propensity for superinfection.
Figures
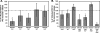
Comment in
-
Peptidoglycan monomer release and Vibrio fischeri.J Bacteriol. 2009 Apr;191(7):1997-9. doi: 10.1128/JB.01801-08. Epub 2009 Jan 16. J Bacteriol. 2009. PMID: 19151142 Free PMC article. No abstract available.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Bose, J. L., U. Kim, W. Bartkowski, R. P. Gunsalus, A. M. Overley, N. L. Lyell, K. L. Visick, and E. V. Stabb. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65538-553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

