Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci
- PMID: 19074389
- PMCID: PMC2632007
- DOI: 10.1128/JB.01058-08
Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci
Erratum in
- J Bacteriol. 2009 May;191(10):3429
Abstract
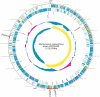
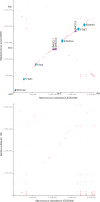
We isolated the methicillin-resistant Macrococcus caseolyticus strain JCSC5402 from animal meat in a supermarket and determined its whole-genome nucleotide sequence. This is the first report on the genome analysis of a macrococcal species that is evolutionarily closely related to the human pathogens Staphylococcus aureus and Bacillus anthracis. The essential biological pathways of M. caseolyticus are similar to those of staphylococci. However, the species has a small chromosome (2.1 MB) and lacks many sugar and amino acid metabolism pathways and a plethora of virulence genes that are present in S. aureus. On the other hand, M. caseolyticus possesses a series of oxidative phosphorylation machineries that are closely related to those in the family Bacillaceae. We also discovered a probable primordial form of a Macrococcus methicillin resistance gene complex, mecIRAm, on one of the eight plasmids harbored by the M. caseolyticus strain. This is the first finding of a plasmid-encoding methicillin resistance gene. Macrococcus is considered to reflect the genome of ancestral bacteria before the speciation of staphylococcal species and may be closely associated with the origin of the methicillin resistance gene complex of the notorious human pathogen methicillin-resistant S. aureus.
Figures



References
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2591819-1827. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, T. Ito, H. Yuzawa, and K. Hiramatsu. 2003. The genome of Staphylococcus aureus, p. 66-153. In D. Al'Aladeen and Hiramatsu (ed.), Staphylococcus aureus: molecular and clinical aspects. Ellis Harwood, London, United Kingdom.
-
- Bae, T., T. Baba, K. Hiramatsu, and O. Schneewind. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 621035-1047. - PubMed
-
- Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226235-240. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

