Effects of spaceflight on murine skeletal muscle gene expression
- PMID: 19074574
- PMCID: PMC2644242
- DOI: 10.1152/japplphysiol.90780.2008
Effects of spaceflight on murine skeletal muscle gene expression
Erratum in
- J Appl Physiol. 2011 Jan;110(1):298
Abstract
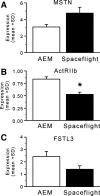
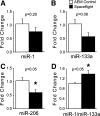
Spaceflight results in a number of adaptations to skeletal muscle, including atrophy and shifts toward faster muscle fiber types. To identify changes in gene expression that may underlie these adaptations, we used both microarray expression analysis and real-time polymerase chain reaction to quantify shifts in mRNA levels in the gastrocnemius from mice flown on the 11-day, 19-h STS-108 shuttle flight and from normal gravity controls. Spaceflight data also were compared with the ground-based unloading model of hindlimb suspension, with one group of pure suspension and one of suspension followed by 3.5 h of reloading to mimic the time between landing and euthanization of the spaceflight mice. Analysis of microarray data revealed that 272 mRNAs were significantly altered by spaceflight, the majority of which displayed similar responses to hindlimb suspension, whereas reloading tended to counteract these responses. Several mRNAs altered by spaceflight were associated with muscle growth, including the phosphatidylinositol 3-kinase regulatory subunit p85alpha, insulin response substrate-1, the forkhead box O1 transcription factor, and MAFbx/atrogin1. Moreover, myostatin mRNA expression tended to increase, whereas mRNA levels of the myostatin inhibitor FSTL3 tended to decrease, in response to spaceflight. In addition, mRNA levels of the slow oxidative fiber-associated transcriptional coactivator peroxisome proliferator-associated receptor (PPAR)-gamma coactivator-1alpha and the transcription factor PPAR-alpha were significantly decreased in spaceflight gastrocnemius. Finally, spaceflight resulted in a significant decrease in levels of the microRNA miR-206. Together these data demonstrate that spaceflight induces significant changes in mRNA expression of genes associated with muscle growth and fiber type.
Figures

References
-
- Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol 292: C188–C199, 2007. - PubMed
-
- Argraves GL, Barth JL, Argraves WS. The MUSC DNA Microarray Database. Bioinformatics 19: 2473–2474, 2003. - PubMed
-
- Bey L, Akunuri N, Zhao P, Hoffman EP, Hamilton DG, Hamilton MT. Patterns of global gene expression in rat skeletal muscle during unloading and low-intensity ambulatory activity. Physiol Genomics 13: 157–167, 2003. - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193, 2003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

