Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL
- PMID: 19075186
- PMCID: PMC2676092
- DOI: 10.1182/blood-2008-08-174748
Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL
Abstract
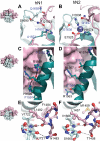
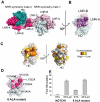
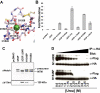
Proteolytic resistance of Notch prior to ligand binding depends on the structural integrity of a negative regulatory region (NRR) of the receptor that immediately precedes the transmembrane segment. The NRR includes the 3 Lin12/Notch repeats and the juxtamembrane heterodimerization domain, the region of Notch1 most frequently mutated in T-cell acute lymphoblastic leukemia lymphoma (T-ALL). Here, we report the x-ray structure of the Notch1 NRR in its autoinhibited conformation. A key feature of the Notch1 structure that maintains its closed conformation is a conserved hydrophobic plug that sterically occludes the metalloprotease cleavage site. Crystal packing interactions involving a highly conserved, exposed face on the third Lin12/Notch repeat suggest that this site may normally be engaged in intermolecular or intramolecular protein-protein interactions. The majority of known T-ALL-associated point mutations map to residues in the hydrophobic interior of the Notch1 NRR. A novel mutation (H1545P), which alters a residue at the crystal-packing interface, leads to ligand-independent increases in signaling in reporter gene assays despite only mild destabilization of the NRR, suggesting that it releases the autoinhibitory clamp on the heterodimerization domain imposed by the Lin12/Notch repeats. The Notch1 NRR structure should facilitate a search for antibodies or compounds that stabilize the autoinhibited conformation.
Figures

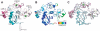


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

