Integrated microfluidic bioprocessor for single-cell gene expression analysis
- PMID: 19075237
- PMCID: PMC2629289
- DOI: 10.1073/pnas.0806355106
Integrated microfluidic bioprocessor for single-cell gene expression analysis
Abstract
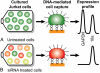
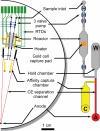
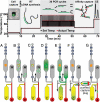
An integrated microdevice is developed for the analysis of gene expression in single cells. The system captures a single cell, transcribes and amplifies the mRNA, and quantitatively analyzes the products of interest. The key components of the microdevice include integrated nanoliter metering pumps, a 200-nL RT-PCR reactor with a single-cell capture pad, and an affinity capture matrix for the purification and concentration of products that is coupled to a microfabricated capillary electrophoresis separation channel for product analysis. Efficient microchip integration of these processes enables the sensitive and quantitative examination of gene expression variation at the single-cell level. This microdevice is used to measure siRNA knockdown of the GAPDH gene in individual Jurkat cells. Single-cell measurements suggests the presence of 2 distinct populations of cells with moderate (approximately 50%) or complete (approximately 0%) silencing. This stochastic variation in gene expression and silencing within single cells is masked by conventional bulk measurements.
Conflict of interest statement
Conflict of interest statement: R.A.M. has a financial interest in Microchip Biotechnologies, Inc., which is commercially developing aspects of the technologies presented here.
Figures

References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. - PubMed
-
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. - PubMed
-
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

