Studies on the paramyxovirus accessory genes by reverse genetics in the Sendai virus-mouse system
- PMID: 19075516
- PMCID: PMC3720547
- DOI: 10.2183/pjab.84.439
Studies on the paramyxovirus accessory genes by reverse genetics in the Sendai virus-mouse system
Abstract
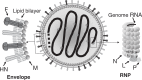
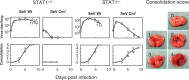
Nucleotide sequencing of the entire genomes was completed in the 1980s for most members of the Paramyxoviridae. It then became a new common task with challenge for researchers in the field to establish a system to recover the virus entirely from cDNA, thereby allowing reverse genetics (free manipulation of the viral genome). Using Sendai virus, we established a system of incomparable virus recovery efficiency early on. This technology was then fully exploited in answering a series of long-held questions. In particular, two accessory genes whose functions had remained enigmatic were demonstrated to encode special functions critical in viral in vivo pathogenesis producing fatal pneumonia in mice, although dispensable in virus replication at the in vitro cellular level. Their in vivo functions were found to counteract the two respective facets of the antiviral state induced by interferons and an interferon regulatory factor 3-dependent but yet unknown effector. These achievements appear to have facilitated a scientific trend where the accessory genes are a focus of active investigation in studies on other paramyxoviruses and opened up a new common ground shared between virology and immunology.
Figures




Similar articles
-
A Single Amino Acid Substitution within the Paramyxovirus Sendai Virus Nucleoprotein Is a Critical Determinant for Production of Interferon-Beta-Inducing Copyback-Type Defective Interfering Genomes.J Virol. 2018 Feb 12;92(5):e02094-17. doi: 10.1128/JVI.02094-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237838 Free PMC article.
-
Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding.Rev Med Virol. 1999 Apr-Jun;9(2):83-99. doi: 10.1002/(sici)1099-1654(199904/06)9:2<83::aid-rmv244>3.0.co;2-5. Rev Med Virol. 1999. PMID: 10386336 Review.
-
Rescue of Sendai Virus from Cloned cDNA.Methods Mol Biol. 2017;1602:103-110. doi: 10.1007/978-1-4939-6964-7_7. Methods Mol Biol. 2017. PMID: 28508216
-
Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence.Nat Commun. 2017 Oct 6;8(1):799. doi: 10.1038/s41467-017-00909-6. Nat Commun. 2017. PMID: 28986577 Free PMC article.
-
Accessory genes of the paramyxoviridae, a large family of nonsegmented negative-strand RNA viruses, as a focus of active investigation by reverse genetics.Curr Top Microbiol Immunol. 2004;283:197-248. doi: 10.1007/978-3-662-06099-5_6. Curr Top Microbiol Immunol. 2004. PMID: 15298171 Review.
Cited by
-
Oncolytic Sendai virus-based virotherapy for cancer: recent advances.Oncolytic Virother. 2015 Oct 1;4:141-7. doi: 10.2147/OV.S66419. eCollection 2015. Oncolytic Virother. 2015. PMID: 27512677 Free PMC article. Review.
-
Inactivated Sendai virus particle upregulates cancer cell expression of intercellular adhesion molecule-1 and enhances natural killer cell sensitivity on cancer cells.Cancer Sci. 2017 Dec;108(12):2333-2341. doi: 10.1111/cas.13408. Epub 2017 Oct 16. Cancer Sci. 2017. PMID: 28945328 Free PMC article.
-
RNase L Amplifies Interferon Signaling by Inducing Protein Kinase R-Mediated Antiviral Stress Granules.J Virol. 2020 Jun 16;94(13):e00205-20. doi: 10.1128/JVI.00205-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295917 Free PMC article.
-
Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein.J Virol. 2012 Jul;86(13):7136-45. doi: 10.1128/JVI.06705-11. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532687 Free PMC article.
References
-
- Lamb, R.A., Collins, P.L., Kolakofsky, D., Maleno, J.A., Nagai, Y. and Oldstone, M.B. (2005) Paramyxoviridae. InVirus Taxonomy: Classification and Nomenclature of Viruses; Eighth Report of the International Committee on Taxonomy of Viruses (eds. Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U. and Ball, L.A.). Academic Press, London, pp. 655–668
-
- Kato, A., Sakai, Y., Shioda, T., Kondo, T., Nakanishi, M. and Nagai, Y. (1996) Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1, 569–579 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

