Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator
- PMID: 19076454
- PMCID: PMC2853241
- DOI: 10.1196/annals.1427.006
Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator
Abstract
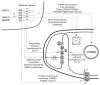
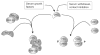
Expression of the respiratory apparatus depends on both nuclear and mitochondrial genes. Although these genes are sequestered in distinct cellular organelles, their transcription relies on nucleus-encoded factors. Certain of these factors are directed to the mitochondria, where they sponsor the bi-directional transcription of mitochondrial DNA. Others act on nuclear genes that encode the majority of the respiratory subunits and many other gene products required for the assembly and function of the respiratory chain. The nuclear respiratory factors, NRF-1 and NRF-2, contribute to the expression of respiratory subunits and mitochondrial transcription factors and thus have been implicated in nucleo-mitochondrial interactions. In addition, coactivators of the PGC-1 family serve as mediators between the environment and the transcriptional machinery governing mitochondrial biogenesis. One family member, peroxisome proliferator-activated receptor gamma coactivator PGC-1-related coactivator (PRC), is an immediate early gene product that is rapidly induced by mitogenic signals in the absence of de novo protein synthesis. Like other PGC-1 family members, PRC binds NRF-1 and activates NRF-1 target genes. In addition, PRC complexes with NRF-2 and HCF-1 (host cell factor-1) in the activation of NRF-2-dependent promoters. HCF-1 functions in cell-cycle progression and has been identified as an NRF-2 coactivator. The association of these factors with PRC is suggestive of a role for the complex in cell growth. Finally, shRNA-mediated knock down of PRC expression results in a complex phenotype that includes the inhibition of respiratory growth on galactose and the loss of respiratory complexes. Thus, PRC may help integrate the expression of the respiratory apparatus with the cell proliferative program.
Conflict of interest statement
The author declares no conflicts of interest.
Figures


Similar articles
-
PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression.J Biol Chem. 2008 May 2;283(18):12102-11. doi: 10.1074/jbc.M710150200. Epub 2008 Mar 14. J Biol Chem. 2008. PMID: 18343819 Free PMC article.
-
Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells.Gene. 2002 Mar 6;286(1):81-9. doi: 10.1016/s0378-1119(01)00809-5. Gene. 2002. PMID: 11943463
-
Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells.Mol Cell Biol. 2001 Jun;21(11):3738-49. doi: 10.1128/MCB.21.11.3738-3749.2001. Mol Cell Biol. 2001. PMID: 11340167 Free PMC article.
-
Nuclear control of respiratory gene expression in mammalian cells.J Cell Biochem. 2006 Mar 1;97(4):673-83. doi: 10.1002/jcb.20743. J Cell Biochem. 2006. PMID: 16329141 Review.
-
Nuclear activators and coactivators in mammalian mitochondrial biogenesis.Biochim Biophys Acta. 2002 Jun 7;1576(1-2):1-14. doi: 10.1016/s0167-4781(02)00343-3. Biochim Biophys Acta. 2002. PMID: 12031478 Review.
Cited by
-
Mitochondrial Dysfunction and Oxidative Stress in Alzheimer's Disease.Front Aging Neurosci. 2021 Feb 18;13:617588. doi: 10.3389/fnagi.2021.617588. eCollection 2021. Front Aging Neurosci. 2021. PMID: 33679375 Free PMC article. Review.
-
Onset and organ specificity of Tk2 deficiency depends on Tk1 down-regulation and transcriptional compensation.Hum Mol Genet. 2011 Jan 1;20(1):155-64. doi: 10.1093/hmg/ddq453. Epub 2010 Oct 11. Hum Mol Genet. 2011. PMID: 20940150 Free PMC article.
-
Changes in the Mitochondria in the Aging Process-Can α-Tocopherol Affect Them?Int J Mol Sci. 2023 Aug 5;24(15):12453. doi: 10.3390/ijms241512453. Int J Mol Sci. 2023. PMID: 37569829 Free PMC article. Review.
-
Mitochondria and PGC-1α in Aging and Age-Associated Diseases.J Aging Res. 2011;2011:810619. doi: 10.4061/2011/810619. Epub 2011 May 5. J Aging Res. 2011. PMID: 21629705 Free PMC article.
-
GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis.Mol Cell Biol. 2014 Sep;34(17):3194-201. doi: 10.1128/MCB.00492-12. Epub 2014 Jun 23. Mol Cell Biol. 2014. PMID: 24958105 Free PMC article.
References
-
- Hatefi Y. The mitochondrial electron transport chain and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. - PubMed
-
- Cannon B, Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol Rev. 2004;84:277–359. - PubMed
-
- Scarpulla RC. Molecular biology of the OXPHOS system. In: Smeitink JAM, Sengers RCA, Trij JMF, editors. Oxidative Phosphorylation in Health and Disease. Landes Bioscience; New York, NY: 2004. pp. 28–42.
-
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: Multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

