Occult hepatitis B infection: an evolutionary scenario
- PMID: 19077239
- PMCID: PMC2637267
- DOI: 10.1186/1743-422X-5-146
Occult hepatitis B infection: an evolutionary scenario
Abstract
Background: Occult or latent hepatitis B virus (HBV) infection is defined as infection with detectable HBV DNA and undetectable surface antigen (HBsAg) in patients' blood. The cause of an overt HBV infection becoming an occult one is unknown. To gain insight into the mechanism of the development of occult infection, we compared the full-length HBV genome from a blood donor carrying an occult infection (d4) with global genotype D genomes.
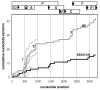
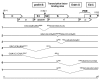
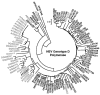
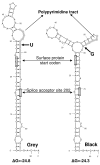
Results: The phylogenetic analysis of polymerase, core and X protein sequences did not distinguish d4 from other genotype D strains. Yet, d4 surface protein formed the evolutionary outgroup relative to all other genotype D strains. Its evolutionary branch was the only one where accumulation of substitutions suggests positive selection (dN/dS = 1.3787). Many of these substitutions accumulated specifically in regions encoding the core/surface protein interface, as revealed in a 3D-modeled protein complex. We identified a novel RNA splicing event (deleting nucleotides 2986-202) that abolishes surface protein gene expression without affecting polymerase, core and X-protein related functions. Genotype D strains differ in their ability to perform this 2986-202 splicing. Strains prone to 2986-202 splicing constitute a separate clade in a phylogenetic tree of genotype D HBVs. A single substitution (G173T) that is associated with clade membership alters the local RNA secondary structure and is proposed to affect splicing efficiency at the 202 acceptor site.
Conclusion: We propose an evolutionary scenario for occult HBV infection, in which 2986-202 splicing generates intracellular virus particles devoid of surface protein, which subsequently accumulates mutations due to relaxation of coding constraints. Such viruses are deficient of autonomous propagation and cannot leave the host cell until it is lysed.
Figures


References
-
- Garfein RS, Bower WA, Loney CM, Hutin YJ, Xia GL, Jawanda J, Groom AV, Nainan OV, Murphy JS, Bell BP. Factors associated with fulminant liver failure during an outbreak among injection drug users with acute hepatitis B. Hepatology. 2004;40:865–873. - PubMed
-
- Gerlich WH, Wagner FF, Chudy M, Harritshoj LH, Lattermann A, Wienzek S, Glebe D, Saniewski M, Schüttler CG, Wend UC, et al. HBsAg Non-Reactive HBV Infection in Blood Donors: Transmission and Pathogenicity. J Med Virol. 2007;79:S32–S36. doi: 10.1002/jmv.20963. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

