Knottin cyclization: impact on structure and dynamics
- PMID: 19077275
- PMCID: PMC2659701
- DOI: 10.1186/1472-6807-8-54
Knottin cyclization: impact on structure and dynamics
Abstract
Background: Present in various species, the knottins (also referred to as inhibitor cystine knots) constitute a group of extremely stable miniproteins with a plethora of biological activities. Owing to their small size and their high stability, knottins are considered as excellent leads or scaffolds in drug design. Two knottin families contain macrocyclic compounds, namely the cyclotides and the squash inhibitors. The cyclotide family nearly exclusively contains head-to-tail cyclized members. On the other hand, the squash family predominantly contains linear members. Head-to-tail cyclization is intuitively expected to improve bioactivities by increasing stability and lowering flexibility as well as sensitivity to proteolytic attack.
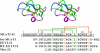
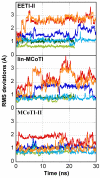
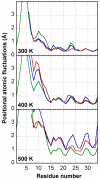
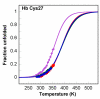
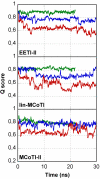
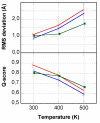
Results: In this paper, we report data on solution structure, thermal stability, and flexibility as inferred from NMR experiments and molecular dynamics simulations of a linear squash inhibitor EETI-II, a circular squash inhibitor MCoTI-II, and a linear analog lin-MCoTI. Strikingly, the head-to-tail linker in cyclic MCoTI-II is by far the most flexible region of all three compounds. Moreover, we show that cyclic and linear squash inhibitors do not display large differences in structure or flexibility in standard conditions, raising the question as to why few squash inhibitors have evolved into cyclic compounds. The simulations revealed however that the cyclization increases resistance to high temperatures by limiting structure unfolding.
Conclusion: In this work, we show that, in contrast to what could have been intuitively expected, cyclization of squash inhibitors does not provide clear stability or flexibility modification. Overall, our results suggest that, for squash inhibitors in standard conditions, the circularization impact might come from incorporation of an additional loop sequence, that can contribute to the miniprotein specificity and affinity, rather than from an increase in conformational rigidity or protein stability. Unfolding simulations showed however that cyclization is a stabilizing factor in strongly denaturing conditions. This information should be useful if one wants to use the squash inhibitor scaffold in drug design.
Figures
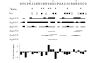

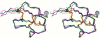
Similar articles
-
Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins.Biochemistry. 2001 Jul 10;40(27):7973-83. doi: 10.1021/bi0106639. Biochemistry. 2001. PMID: 11434766
-
Squash inhibitors: from structural motifs to macrocyclic knottins.Curr Protein Pept Sci. 2004 Oct;5(5):341-349. doi: 10.2174/1389203043379477. Curr Protein Pept Sci. 2004. PMID: 15551519 Review.
-
Discovery and characterization of a linear cyclotide from Viola odorata: implications for the processing of circular proteins.J Mol Biol. 2006 Apr 14;357(5):1522-35. doi: 10.1016/j.jmb.2006.01.051. Epub 2006 Feb 2. J Mol Biol. 2006. PMID: 16488428
-
Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor.J Biol Chem. 2013 Dec 13;288(50):36141-8. doi: 10.1074/jbc.M113.528240. Epub 2013 Oct 29. J Biol Chem. 2013. PMID: 24169696 Free PMC article.
-
The cyclotide family of circular miniproteins: nature's combinatorial peptide template.Biopolymers. 2006;84(3):250-66. doi: 10.1002/bip.20451. Biopolymers. 2006. PMID: 16440288 Review.
Cited by
-
High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli.Microb Cell Fact. 2013 Apr 22;12:37. doi: 10.1186/1475-2859-12-37. Microb Cell Fact. 2013. PMID: 23607455 Free PMC article.
-
Characterization Of Blood-Brain Barrier Crossing And Tumor Homing Peptides By Molecular Dynamics Simulations.Int J Nanomedicine. 2019 Dec 30;14:10123-10136. doi: 10.2147/IJN.S225793. eCollection 2019. Int J Nanomedicine. 2019. PMID: 31920308 Free PMC article.
-
Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the Fabaceae family.J Biol Chem. 2011 Jul 8;286(27):24275-87. doi: 10.1074/jbc.M111.229922. Epub 2011 May 19. J Biol Chem. 2011. PMID: 21596752 Free PMC article.
-
Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I.Chembiochem. 2009 Nov 2;10(16):2663-70. doi: 10.1002/cbic.200900534. Chembiochem. 2009. PMID: 19780078 Free PMC article. No abstract available.
-
Peptide-based protease inhibitors from plants.Drug Discov Today. 2019 Sep;24(9):1877-1889. doi: 10.1016/j.drudis.2019.05.026. Epub 2019 Jun 3. Drug Discov Today. 2019. PMID: 31170506 Free PMC article. Review.
References
-
- Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, Ha PT, Hernandez JF, Le-Nguyen D. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5:341–349. - PubMed
-
- Werle M, Schmitz T, Huang HL, Wentzel A, Kolmar H, Bernkop-Schnurch A. The potential of cystine-knot microproteins as novel pharmacophoric scaffolds in oral peptide drug delivery. J Drug Target. 2006;14:137–146. - PubMed
-
- Reiss S, Sieber M, Oberle V, Wentzel A, Spangenberg P, Claus R, Kolmar H, Losche W. Inhibition of platelet aggregation by grafting RGD and KGD sequences on the structural scaffold of small disulfide-rich proteins. Platelets. 2006;17:153–157. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

