Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence
- PMID: 19077304
- PMCID: PMC2646275
- DOI: 10.1186/gb-2008-9-12-r171
Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence
Abstract
Background: The malaria parasite Plasmodium falciparum exhibits abundant genetic diversity, and this diversity is key to its success as a pathogen. Previous efforts to study genetic diversity in P. falciparum have begun to elucidate the demographic history of the species, as well as patterns of population structure and patterns of linkage disequilibrium within its genome. Such studies will be greatly enhanced by new genomic tools and recent large-scale efforts to map genomic variation. To that end, we have developed a high throughput single nucleotide polymorphism (SNP) genotyping platform for P. falciparum.
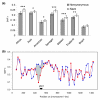
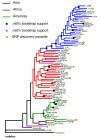
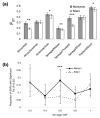
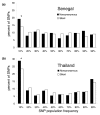
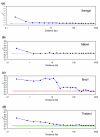
Results: Using an Affymetrix 3,000 SNP assay array, we found roughly half the assays (1,638) yielded high quality, 100% accurate genotyping calls for both major and minor SNP alleles. Genotype data from 76 global isolates confirm significant genetic differentiation among continental populations and varying levels of SNP diversity and linkage disequilibrium according to geographic location and local epidemiological factors. We further discovered that nonsynonymous and silent (synonymous or noncoding) SNPs differ with respect to within-population diversity, inter-population differentiation, and the degree to which allele frequencies are correlated between populations.
Conclusions: The distinct population profile of nonsynonymous variants indicates that natural selection has a significant influence on genomic diversity in P. falciparum, and that many of these changes may reflect functional variants deserving of follow-up study. Our analysis demonstrates the potential for new high-throughput genotyping technologies to enhance studies of population structure, natural selection, and ultimately enable genome-wide association studies in P. falciparum to find genes underlying key phenotypic traits.
Figures
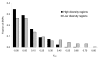

References
-
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. - DOI - PubMed
-
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

