UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications
- PMID: 19077538
- PMCID: PMC2661099
- DOI: 10.4161/epi.4.1.7370
UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications
Abstract
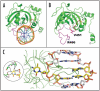
Cytosine methylation in DNA is a major epigenetic signal, and plays a central role in propagating chromatin status during cell division. However the mechanistic links between DNA methylation and histone methylation are poorly understood. A multi-domain protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is required for DNA CpG maintenance methylation at replication forks, and mouse UHRF1-null cells show enhanced susceptibility to DNA replication arrest and DNA damaging agents. Recent data demonstrated that the SET and RING associated (SRA) domain of UHRF1 binds hemimethylated CpG and flips 5-methylcytosine out of the DNA helix, whereas its tandom tudor domain and PHD domain bind the tail of histone H3 in a highly methylation sensitive manner. We hypothesize that UHRF1 brings the two components (histones and DNA) carrying appropriate markers (on the tails of H3 and hemimethylated CpG sites) ready to be assembled into a nucleosome after replication.
Figures





Similar articles
-
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix.Nature. 2008 Oct 9;455(7214):826-9. doi: 10.1038/nature07280. Epub 2008 Sep 3. Nature. 2008. PMID: 18772888 Free PMC article.
-
Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein.J Biol Chem. 2013 Jan 11;288(2):1329-39. doi: 10.1074/jbc.M112.415398. Epub 2012 Nov 16. J Biol Chem. 2013. PMID: 23161542 Free PMC article.
-
The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism.J Biol Chem. 2014 Feb 14;289(7):4106-15. doi: 10.1074/jbc.M113.528893. Epub 2013 Dec 24. J Biol Chem. 2014. PMID: 24368767 Free PMC article.
-
Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis.Biochem Pharmacol. 2013 Dec 15;86(12):1643-9. doi: 10.1016/j.bcp.2013.10.002. Epub 2013 Oct 14. Biochem Pharmacol. 2013. PMID: 24134914 Review.
-
Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties.J Exp Clin Cancer Res. 2011 Apr 15;30(1):41. doi: 10.1186/1756-9966-30-41. J Exp Clin Cancer Res. 2011. PMID: 21496237 Free PMC article. Review.
Cited by
-
Role of DNMTs in the Brain.Adv Exp Med Biol. 2022;1389:363-394. doi: 10.1007/978-3-031-11454-0_15. Adv Exp Med Biol. 2022. PMID: 36350518 Review.
-
Understanding the structural and dynamic consequences of DNA epigenetic modifications: computational insights into cytosine methylation and hydroxymethylation.Epigenetics. 2014 Dec;9(12):1604-12. doi: 10.4161/15592294.2014.988043. Epigenetics. 2014. PMID: 25625845 Free PMC article.
-
Targeting hypoxic tumor microenvironment in pancreatic cancer.J Hematol Oncol. 2021 Jan 13;14(1):14. doi: 10.1186/s13045-020-01030-w. J Hematol Oncol. 2021. PMID: 33436044 Free PMC article. Review.
-
Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and RING finger domains 1 in breast cancer patients.Cancer Sci. 2013 Feb;104(2):194-9. doi: 10.1111/cas.12052. Epub 2012 Dec 13. Cancer Sci. 2013. PMID: 23107467 Free PMC article.
-
The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity.Nucleic Acids Res. 2021 Jun 21;49(11):6053-6068. doi: 10.1093/nar/gkab293. Nucleic Acids Res. 2021. PMID: 33939809 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
