Circadian gene expression is resilient to large fluctuations in overall transcription rates
- PMID: 19078963
- PMCID: PMC2634731
- DOI: 10.1038/emboj.2008.262
Circadian gene expression is resilient to large fluctuations in overall transcription rates
Abstract
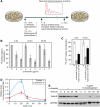
Mammalian circadian oscillators are considered to rely on transcription/translation feedback loops in clock gene expression. The major and essential loop involves the autorepression of cryptochrome (Cry1, Cry2) and period (Per1, Per2) genes. The rhythm-generating circuitry is functional in most cell types, including cultured fibroblasts. Using this system, we show that significant reduction in RNA polymerase II-dependent transcription did not abolish circadian oscillations, but surprisingly accelerated them. A similar period shortening was observed at reduced incubation temperatures in wild-type mouse fibroblasts, but not in cells lacking Per1. Our data suggest that mammalian circadian oscillators are resilient to large fluctuations in general transcription rates and temperature, and that PER1 has an important function in transcription and temperature compensation.
Figures
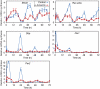
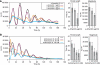

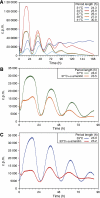
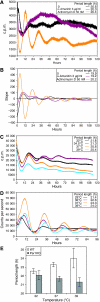
 with parameters s=0.064 [A]/h, dA=0.32/h, f=5.8 [A]/h, e=1.6 [R]−1/h, k=0.64 [A]−1/h and dR=0.15/h. Here, transcription and translation processes are taken together. The model has an infinite period bifurcation near ρ=0.27 and a Hopf bifurcation at ρ=2.2. The period shortens with increasing ρ around ρ=1. (C) The 16-dimensional mammalian model by Leloup and Goldbeter. With its standard parameters, this model has a very narrow range of oscillation around ρ=1 within ∼10% of variation of the transcription rates in either direction (Hopf bif urcations at ρ=0.94 and ρ=1.14). Furthermore, the model shows a cyclic fold in a narrow window around ρ=1.1 with coexistence of two stable (plain) and one unstable limit cycle (dashed line). The period lengthens with increasing ρ around ρ=1. (D) The 74-dimensional mammalian Forger–Peskin model. Here, the model has both Per1 and Per2 genes, and it is thus also possible to simulate the Per1 knockout phenotypes (dashed line). In contrast to the experimental data, the Per1 mutant oscillators display slightly longer periods. Transcription can be reduced almost to zero while keeping oscillations. The period shortens with increasing ρ around ρ=1. Oscillation amplitudes for the same four models are shown in Supplementary Figure 4.
with parameters s=0.064 [A]/h, dA=0.32/h, f=5.8 [A]/h, e=1.6 [R]−1/h, k=0.64 [A]−1/h and dR=0.15/h. Here, transcription and translation processes are taken together. The model has an infinite period bifurcation near ρ=0.27 and a Hopf bifurcation at ρ=2.2. The period shortens with increasing ρ around ρ=1. (C) The 16-dimensional mammalian model by Leloup and Goldbeter. With its standard parameters, this model has a very narrow range of oscillation around ρ=1 within ∼10% of variation of the transcription rates in either direction (Hopf bif urcations at ρ=0.94 and ρ=1.14). Furthermore, the model shows a cyclic fold in a narrow window around ρ=1.1 with coexistence of two stable (plain) and one unstable limit cycle (dashed line). The period lengthens with increasing ρ around ρ=1. (D) The 74-dimensional mammalian Forger–Peskin model. Here, the model has both Per1 and Per2 genes, and it is thus also possible to simulate the Per1 knockout phenotypes (dashed line). In contrast to the experimental data, the Per1 mutant oscillators display slightly longer periods. Transcription can be reduced almost to zero while keeping oscillations. The period shortens with increasing ρ around ρ=1. Oscillation amplitudes for the same four models are shown in Supplementary Figure 4.Comment in
-
Circadian clocks can take a few transcriptional knocks.EMBO J. 2009 Jan 21;28(2):84-5. doi: 10.1038/emboj.2008.272. EMBO J. 2009. PMID: 19158661 Free PMC article.
References
-
- Adolph S, Brusselbach S, Muller R (1993) Inhibition of transcription blocks cell cycle progression of NIH3T3 fibroblasts specifically in G1. J Cell Sci 105 (Part 1): 113–122 - PubMed
-
- Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y, Nakao M, Saya H (2005) Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J Biol Chem 280: 19166–19176 - PubMed
-
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 - PubMed
-
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

