Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites
- PMID: 19078966
- PMCID: PMC2634730
- DOI: 10.1038/emboj.2008.257
Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites
Abstract
The function of histone modifications in initiating and regulating the chromosomal events of the meiotic prophase remains poorly understood. In Saccharomyces cerevisiae, we examined the genome-wide localization of histone H3 lysine 4 trimethylation (H3K4me3) along meiosis and its relationship to gene expression and position of the programmed double-strand breaks (DSBs) that initiate interhomologue recombination, essential to yield viable haploid gametes. We find that the level of H3K4me3 is constitutively higher close to DSB sites, independently of local gene expression levels. Without Set1, the H3K4 methylase, 84% of the DSB sites exhibit a severely reduced DSB frequency, the reduction being quantitatively correlated with the local level of H3K4me3 in wild-type cells. Further, we show that this differential histone mark is already established in vegetative cells, being higher in DSB-prone regions than in regions with no or little DSB. Taken together, our results demonstrate that H3K4me3 is a prominent and preexisting mark of active meiotic recombination initiation sites. Novel perspectives to dissect the various layers of the controls of meiotic DSB formation are discussed.
Figures
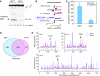
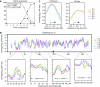


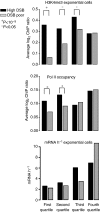


Comment in
-
Histone methylation sets the stage for meiotic DNA breaks.EMBO J. 2009 Jan 21;28(2):81-3. doi: 10.1038/emboj.2008.277. EMBO J. 2009. PMID: 19158660 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 - PubMed
-
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

