Histone H3 tail clipping regulates gene expression
- PMID: 19079264
- PMCID: PMC3350865
- DOI: 10.1038/nsmb.1534
Histone H3 tail clipping regulates gene expression
Abstract
Induction of gene expression in yeast and human cells involves changes in the histone modifications associated with promoters. Here we identify a histone H3 endopeptidase activity in Saccharomyces cerevisiae that may regulate these events. The endopeptidase cleaves H3 after Ala21, generating a histone that lacks the first 21 residues and shows a preference for H3 tails carrying repressive modifications. In vivo, the H3 N terminus is clipped, specifically within the promoters of genes following the induction of transcription. H3 clipping precedes the process of histone eviction seen when genes become fully active. A truncated H3 product is not generated in yeast carrying a mutation of the endopeptidase recognition site (H3 Q19A L20A) and gene induction is defective in these cells. These findings identify clipping of H3 tails as a previously uncharacterized modification of promoter-bound nucleosomes, which may result in the localized clearing of repressive signals during the induction of gene expression.
Figures

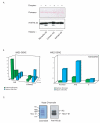
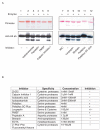

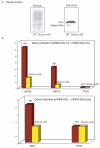
References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;8:900–905. - PubMed
-
- Schermer UJ, Korber P, H^rz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–85. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

