The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex
- PMID: 19079572
- PMCID: PMC2586088
- DOI: 10.1371/journal.pgen.1000302
The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex
Abstract
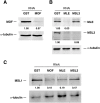
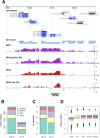
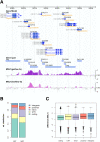
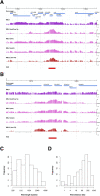
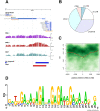
Dosage compensation in male Drosophila relies on the X chromosome-specific recruitment of a chromatin-modifying machinery, the dosage compensation complex (DCC). The principles that assure selective targeting of the DCC are unknown. According to a prevalent model, X chromosome targeting is initiated by recruitment of the DCC core components, MSL1 and MSL2, to a limited number of so-called "high-affinity sites" (HAS). Only very few such sites are known at the DNA sequence level, which has precluded the definition of DCC targeting principles. Combining RNA interference against DCC subunits, limited crosslinking, and chromatin immunoprecipitation coupled to probing high-resolution DNA microarrays, we identified a set of 131 HAS for MSL1 and MSL2 and confirmed their properties by various means. The HAS sites are distributed all over the X chromosome and are functionally important, since the extent of dosage compensation of a given gene and its proximity to a HAS are positively correlated. The sites are mainly located on non-coding parts of genes and predominantly map to regions that are devoid of nucleosomes. In contrast, the bulk of DCC binding is in coding regions and is marked by histone H3K36 methylation. Within the HAS, repetitive DNA sequences mainly based on GA and CA dinucleotides are enriched. Interestingly, DCC subcomplexes bind a small number of autosomal locations with similar features.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex.Genes Dev. 2006 Apr 1;20(7):858-70. doi: 10.1101/gad.1399406. Epub 2006 Mar 17. Genes Dev. 2006. PMID: 16547172 Free PMC article.
-
The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome.Nucleic Acids Res. 2010 Jun;38(10):3209-21. doi: 10.1093/nar/gkq026. Epub 2010 Feb 5. Nucleic Acids Res. 2010. PMID: 20139418 Free PMC article.
-
Divergent evolution toward sex chromosome-specific gene regulation in Drosophila.Genes Dev. 2021 Jul 1;35(13-14):1055-1070. doi: 10.1101/gad.348411.121. Epub 2021 Jun 17. Genes Dev. 2021. PMID: 34140353 Free PMC article.
-
X-Chromosome dosage compensation.WormBook. 2005 Jun 25:1-14. doi: 10.1895/wormbook.1.8.1. WormBook. 2005. PMID: 18050416 Free PMC article. Review.
-
Dosage compensation, the origin and the afterlife of sex chromosomes.Chromosome Res. 2006;14(4):417-31. doi: 10.1007/s10577-006-1064-3. Chromosome Res. 2006. PMID: 16821137 Review.
Cited by
-
Mechanisms of x chromosome dosage compensation.J Genomics. 2015 Jan 1;3:1-19. doi: 10.7150/jgen.10404. eCollection 2015. J Genomics. 2015. PMID: 25628761 Free PMC article. Review.
-
Evidence for sequence biases associated with patterns of histone methylation.BMC Genomics. 2012 Aug 2;13:367. doi: 10.1186/1471-2164-13-367. BMC Genomics. 2012. PMID: 22857523 Free PMC article.
-
Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription.Nat Rev Genet. 2012 Jan 18;13(2):123-34. doi: 10.1038/nrg3124. Nat Rev Genet. 2012. PMID: 22251873 Review.
-
Proximity ligation assays of protein and RNA interactions in the male-specific lethal complex on Drosophila melanogaster polytene chromosomes.Chromosoma. 2015 Sep;124(3):385-95. doi: 10.1007/s00412-015-0509-x. Epub 2015 Feb 19. Chromosoma. 2015. PMID: 25694028 Free PMC article.
-
JASPer controls interphase histone H3S10 phosphorylation by chromosomal kinase JIL-1 in Drosophila.Nat Commun. 2019 Nov 25;10(1):5343. doi: 10.1038/s41467-019-13174-6. Nat Commun. 2019. PMID: 31767855 Free PMC article.
References
-
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. - PubMed
-
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. - PubMed
-
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

