Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction
- PMID: 19079580
- PMCID: PMC2587916
- DOI: 10.1371/journal.pgen.1000301
Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction
Abstract
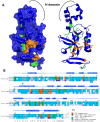
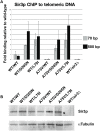
The previously identified LRS (Loss of rDNA Silencing) domain of the nucleosome is critically important for silencing at both ribosomal DNA and telomeres. To understand the function of the LRS surface in silencing, we performed an EMS mutagenesis screen to identify suppressors of the H3 A75V LRS allele. We identified dominant and recessive mutations in histones H3, H4, and dominant mutations in the BAH (Bromo Adjacent Homology) domain of SIR3. We further characterized a surface of Sir3p critical for silencing via the LRS surface. We found that all alleles of the SIR3 BAH domain were able to 1) generally suppress the loss of telomeric silencing of LRS alleles, but 2) could not suppress SIN (Swi/Snf Independent) alleles or 3) could not suppress the telomeric silencing defect of H4 tail alleles. Moreover, we noticed a complementary trend in the electrostatic changes resulting from most of the histone mutations that gain or lose silencing and the suppressor alleles isolated in SIR3, and the genes for histones H3 and H4. Mutations in H3 and H4 genes that lose silencing tend to make the LRS surface more electronegative, whereas mutations that increase silencing make it less electronegative. Conversely, suppressors of LRS alleles in either SIR3, histone H3, or H4 also tend to make their respective surfaces less electronegative. Our results provide genetic evidence for recent data suggesting that the Sir3p BAH domain directly binds the LRS domain. Based on these findings, we propose an electrostatic model for how an extensive surface on the Sir3p BAH domain may regulate docking onto the LRS surface.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes.Mol Cell Biol. 2009 May;29(10):2532-45. doi: 10.1128/MCB.01682-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273586 Free PMC article.
-
A region of the nucleosome required for multiple types of transcriptional silencing in Saccharomyces cerevisiae.Genetics. 2011 Jul;188(3):535-48. doi: 10.1534/genetics.111.129197. Epub 2011 May 5. Genetics. 2011. PMID: 21546544 Free PMC article.
-
Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.Proc Natl Acad Sci U S A. 2013 May 21;110(21):8495-500. doi: 10.1073/pnas.1300126110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650358 Free PMC article.
-
Silent information regulator 3: the Goldilocks of the silencing complex.Genes Dev. 2010 Jan 15;24(2):115-22. doi: 10.1101/gad.1865510. Genes Dev. 2010. PMID: 20080949 Free PMC article. Review.
-
Silent chromatin in yeast: an orchestrated medley featuring Sir3p [corrected].Bioessays. 1998 Jan;20(1):30-40. doi: 10.1002/(SICI)1521-1878(199801)20:1<30::AID-BIES6>3.0.CO;2-W. Bioessays. 1998. PMID: 9504045 Review.
Cited by
-
Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes.Mol Cell Biol. 2009 May;29(10):2532-45. doi: 10.1128/MCB.01682-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273586 Free PMC article.
-
Yeast heterochromatin regulators Sir2 and Sir3 act directly at euchromatic DNA replication origins.PLoS Genet. 2018 May 24;14(5):e1007418. doi: 10.1371/journal.pgen.1007418. eCollection 2018 May. PLoS Genet. 2018. PMID: 29795547 Free PMC article.
-
Structure, expression and functions of MTA genes.Gene. 2016 May 15;582(2):112-21. doi: 10.1016/j.gene.2016.02.012. Epub 2016 Feb 9. Gene. 2016. PMID: 26869315 Free PMC article. Review.
-
A dual role of H4K16 acetylation in the establishment of yeast silent chromatin.EMBO J. 2011 Jun 10;30(13):2610-21. doi: 10.1038/emboj.2011.170. EMBO J. 2011. PMID: 21666601 Free PMC article.
-
Distinguishing between recruitment and spread of silent chromatin structures in Saccharomyces cerevisiae.Elife. 2022 Jan 24;11:e75653. doi: 10.7554/eLife.75653. Elife. 2022. PMID: 35073254 Free PMC article.
References
-
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat Genet. 2002;32:273–279. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

